Oxidative Stress Response Biomarkers of Ovarian Cancer Based on Single-Cell and Bulk RNA Sequencing
- PMID: 36743693
- PMCID: PMC9897923
- DOI: 10.1155/2023/1261039
Oxidative Stress Response Biomarkers of Ovarian Cancer Based on Single-Cell and Bulk RNA Sequencing
Abstract
Background: The occurrence and development of ovarian cancer (OV) are significantly influenced by increased levels of oxidative stress (OS) byproducts and the lack of an antioxidant stress repair system. Hence, it is necessary to explore the markers related to OS in OV, which can aid in predicting the prognosis and immunotherapeutic response in patients with OV.
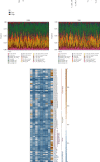
Methods: The single-cell RNA-sequencing (scRNA-seq) dataset GSE146026 was retrieved from the Gene Expression Omnibus (GEO) database, and Bulk RNA-seq data were obtained from TCGA and GTEx databases. The Seurat R package and SingleR package were used to analyze scRNA-seq and to identify OS response-related clusters based on ROS markers. The "limma" R package was used to identify the differentially expressed genes (DEGs) between normal and ovarian samples. The risk model was constructed using the least absolute shrinkage and selection operator (LASSO) regression analysis. The immune cell infiltration, genomic mutation, and drug sensitivity of the model were analyzed using the CIBERSORT algorithm, the "maftools," and the "pRRophetic" R packages, respectively.
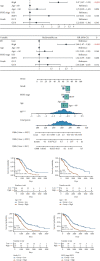
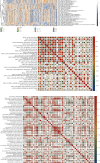
Results: Based on scRNA-seq data, we identified 12 clusters; OS response-related genes had the strongest specificity for cluster 12. A total of 151 genes were identified from 2928 DEGs to be significantly correlated with OS response. Finally, nine prognostic genes were used to construct the risk score (RS) model. The risk score model was an independent prognostic factor for OV. The gene mutation frequency and tumor immune microenvironment in the high- and low-risk score groups were significantly different. The value of the risk score model in predicting immunotherapeutic outcomes was confirmed.
Conclusions: OS response-related RS model could predict the prognosis and immune responses in patients with OV and provide new strategies for cancer treatment.
Copyright © 2023 Mingjun Zheng et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

