8-HQA adjusts the number and diversity of bacteria in the gut microbiome of Spodoptera littoralis
- PMID: 36744087
- PMCID: PMC9891463
- DOI: 10.3389/fmicb.2023.1075557
8-HQA adjusts the number and diversity of bacteria in the gut microbiome of Spodoptera littoralis
Abstract
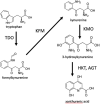
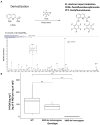
Quinolinic carboxylic acids are known for their metal ion chelating properties in insects, plants and bacteria. The larval stages of the lepidopteran pest, Spodoptera littoralis, produce 8-hydroxyquinoline-2-carboxylic acid (8-HQA) in high concentrations from tryptophan in the diet. At the same time, the larval midgut is known to harbor a bacterial population. The motivation behind the work was to investigate whether 8-HQA is controlling the bacterial community in the gut by regulating the concentration of metal ions. Knocking out the gene for kynurenine 3-monooxygenase (KMO) in the insect using CRISPR/Cas9 eliminated production of 8-HQA and significantly increased bacterial numbers and diversity in the larval midgut. Adding 8-HQA to the diet of knockout larvae caused a dose-dependent reduction of bacterial numbers with minimal effects on diversity. Enterococcus mundtii dominates the community in all treatments, probably due to its highly efficient iron uptake system and production of the colicin, mundticin. Thus host factors and bacterial properties interact to determine patterns of diversity and abundance in the insect midgut.
Keywords: 8-hydroxyquinoline-2-carboxylic acid; Enterococcus mundtii; Spodoptera littoralis; kynurenine 3-monooxygenase; microbiome.
Copyright © 2023 Mazumdar, Hänniger, Shukla, Murali, Bartram, Heckel and Boland.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Biosynthesis of 8-hydroxyquinoline-2-carboxylic acid, an iron chelator from the gut of the lepidopteran Spodoptera littoralis.Org Biomol Chem. 2015 Jan 7;13(1):178-84. doi: 10.1039/c4ob01857e. Org Biomol Chem. 2015. PMID: 25356857
-
Transcriptomics Reveal the Survival Strategies of Enterococcus mundtii in the Gut of Spodoptera littoralis.J Chem Ecol. 2021 Feb;47(2):227-241. doi: 10.1007/s10886-021-01246-1. Epub 2021 Jan 18. J Chem Ecol. 2021. PMID: 33459999
-
Colonization of the Intestinal Tract of the Polyphagous Pest Spodoptera littoralis with the GFP-Tagged Indigenous Gut Bacterium Enterococcus mundtii.Front Microbiol. 2016 Jun 14;7:928. doi: 10.3389/fmicb.2016.00928. eCollection 2016. Front Microbiol. 2016. PMID: 27379058 Free PMC article.
-
Impact of transgenerational host switch on gut bacterial assemblage in generalist pest, Spodoptera littoralis (Lepidoptera: Noctuidae).Front Microbiol. 2023 Jul 13;14:1172601. doi: 10.3389/fmicb.2023.1172601. eCollection 2023. Front Microbiol. 2023. PMID: 37520373 Free PMC article.
-
Native mid-gut bacterial community increases resistance to nucleopolyhedrovirus in the cotton leafworm.Pestic Biochem Physiol. 2025 Aug;212:106462. doi: 10.1016/j.pestbp.2025.106462. Epub 2025 May 22. Pestic Biochem Physiol. 2025. PMID: 40500070
Cited by
-
Comparative Genomics of Pesticide-Degrading Enterococcus Symbionts of Spodoptera frugiperda (Lepidoptera: Noctuidae) Leads to the Identification of Two New Species and the Reappraisal of Insect-Associated Enterococcus Species.Microb Ecol. 2023 Nov;86(4):2583-2605. doi: 10.1007/s00248-023-02264-0. Epub 2023 Jul 11. Microb Ecol. 2023. PMID: 37433981
References
-
- Apprill A., Mcnally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753 - DOI
-
- Beals E. W. (1984). Bray-Curtis ordination: an effective strategy for analysis of multivariate ecological data. Adv. Ecol. Res. 14, 1–55. doi: 10.1016/S0065-2504(08)60168-3 - DOI
-
- Berenbaum M. (1980). Adaptive significance of midgut pH in larval Lepidoptera. Am. Nat. 115, 138–146. doi: 10.1086/283551 - DOI
LinkOut - more resources
Full Text Sources

