The tRNA identity landscape for aminoacylation and beyond
- PMID: 36744444
- PMCID: PMC9976931
- DOI: 10.1093/nar/gkad007
The tRNA identity landscape for aminoacylation and beyond
Abstract
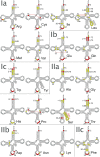
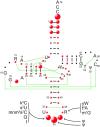
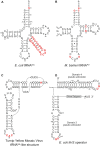
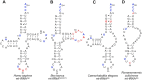
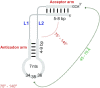
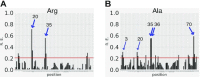
tRNAs are key partners in ribosome-dependent protein synthesis. This process is highly dependent on the fidelity of tRNA aminoacylation by aminoacyl-tRNA synthetases and relies primarily on sets of identities within tRNA molecules composed of determinants and antideterminants preventing mischarging by non-cognate synthetases. Such identity sets were discovered in the tRNAs of a few model organisms, and their properties were generalized as universal identity rules. Since then, the panel of identity elements governing the accuracy of tRNA aminoacylation has expanded considerably, but the increasing number of reported functional idiosyncrasies has led to some confusion. In parallel, the description of other processes involving tRNAs, often well beyond aminoacylation, has progressed considerably, greatly expanding their interactome and uncovering multiple novel identities on the same tRNA molecule. This review highlights key findings on the mechanistics and evolution of tRNA and tRNA-like identities. In addition, new methods and their results for searching sets of multiple identities on a single tRNA are discussed. Taken together, this knowledge shows that a comprehensive understanding of the functional role of individual and collective nucleotide identity sets in tRNA molecules is needed for medical, biotechnological and other applications.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Rak R., Dahan O., Pilpel Y.. Repertoires of tRNAs: the couplers of genomics and proteomics. Annu. Rev. Cell Dev. Biol. 2018; 34:239–264. - PubMed
-
- Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018; 19:45–58. - PubMed
-
- Beuning P.J., Musier-Forsyth K.. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999; 52:1–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

