The isotope distribution: A rose with thorns
- PMID: 36744702
- PMCID: PMC11624904
- DOI: 10.1002/mas.21820
The isotope distribution: A rose with thorns
Abstract
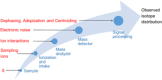
The isotope distribution, which reflects the number and probabilities of occurrence of different isotopologues of a molecule, can be theoretically calculated. With the current generation of (ultra)-high-resolution mass spectrometers, the isotope distribution of molecules can be measured with high sensitivity, resolution, and mass accuracy. However, the observed isotope distribution can differ substantially from the expected isotope distribution. Although differences between the observed and expected isotope distribution can complicate the analysis and interpretation of mass spectral data, they can be helpful in a number of specific applications. These applications include, yet are not limited to, the identification of peptides in proteomics, elucidation of the elemental composition of small organic molecules and metabolites, as well as wading through peaks in mass spectra of complex bioorganic mixtures such as petroleum and humus. In this review, we give a nonexhaustive overview of factors that have an impact on the observed isotope distribution, such as elemental isotope deviations, ion sampling, ion interactions, electronic noise and dephasing, centroiding, and apodization. These factors occur at different stages of obtaining the isotope distribution: during the collection of the sample, during the ionization and intake of a molecule in a mass spectrometer, during the mass separation and detection of ionized molecules, and during signal processing.
Keywords: (ultra)‐high‐resolution mass spectrometry; coalescence; data processing; ion trap; isotope distribution.
© 2023 The Authors. Mass Spectrometry Reviews published by John Wiley & Sons Ltd.
Figures









References
-
- Aebersold R, Mann M. 2003. Mass spectrometry‐based proteomics. Nature. 422: 198‐207. - PubMed
-
- Allwood JW, Parker D, Beckmann M, Draper J, Goodacre R. 2011. Fourier transform ion cyclotron resonance mass spectrometry for plant metabolite profiling and metabolite identification. In: Hardy N., Hall R. (eds) Plant Metabolomics. Methods in Molecular Biology (Methods and Protocols), Vol. 860. pp 157‐176. Humana Press. - PubMed
-
- Beavis RC. 1993. Chemical mass of carbon in proteins. Anal Chem. 65: 496‐497.
Publication types
LinkOut - more resources
Full Text Sources

