A transcriptome atlas of leg muscles from healthy human volunteers reveals molecular and cellular signatures associated with muscle location
- PMID: 36744868
- PMCID: PMC9988256
- DOI: 10.7554/eLife.80500
A transcriptome atlas of leg muscles from healthy human volunteers reveals molecular and cellular signatures associated with muscle location
Abstract
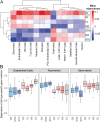
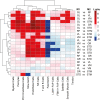
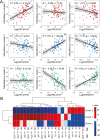
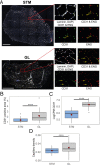
Skeletal muscles support the stability and mobility of the skeleton but differ in biomechanical properties and physiological functions. The intrinsic factors that regulate muscle-specific characteristics are poorly understood. To study these, we constructed a large atlas of RNA-seq profiles from six leg muscles and two locations from one muscle, using biopsies from 20 healthy young males. We identified differential expression patterns and cellular composition across the seven tissues using three bioinformatics approaches confirmed by large-scale newly developed quantitative immune-histology procedures. With all three procedures, the muscle samples clustered into three groups congruent with their anatomical location. Concomitant with genes marking oxidative metabolism, genes marking fast- or slow-twitch myofibers differed between the three groups. The groups of muscles with higher expression of slow-twitch genes were enriched in endothelial cells and showed higher capillary content. In addition, expression profiles of Homeobox (HOX) transcription factors differed between the three groups and were confirmed by spatial RNA hybridization. We created an open-source graphical interface to explore and visualize the leg muscle atlas (https://tabbassidaloii.shinyapps.io/muscleAtlasShinyApp/). Our study reveals the molecular specialization of human leg muscles, and provides a novel resource to study muscle-specific molecular features, which could be linked with (patho)physiological processes.
Keywords: chromosomes; gene expression; human; image analysis; muscle fiber; neuroscience; transcription factors.
© 2023, Abbassi-Daloii et al.
Conflict of interest statement
TA, Se, LV, TV, DC, HM, DM, Ev, P', HK, VR No competing interests declared
Figures











Update of
References
-
- Abbassi-Daloii T. HumanMuscleTranscriptomeAtlasAnalyses. swh:1:rev:d09eff958b768ded8e39ad6a312063504190bd98Software Heritage. 2023 https://archive.softwareheritage.org/swh:1:dir:4b3aa1c7cf4367c3fcd8ac827...
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

