CpH methylome analysis in human cortical neurons identifies novel gene pathways and drug targets for opioid use disorder
- PMID: 36745154
- PMCID: PMC9892724
- DOI: 10.3389/fpsyt.2022.1078894
CpH methylome analysis in human cortical neurons identifies novel gene pathways and drug targets for opioid use disorder
Abstract
Introduction: DNA methylation (DNAm), an epigenetic mechanism, has been associated with opioid use disorder (OUD) in preclinical and human studies. However, most of the studies have focused on DNAm at CpG sites. DNAm at non-CpG sites (mCpHs, where H indicates A, T, or C) has been recently shown to have a role in gene regulation and to be highly abundant in neurons. However, its role in OUD is unknown. This work aims to evaluate mCpHs in the human postmortem orbital frontal cortex (OFC) in the context of OUD.
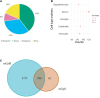
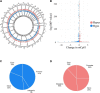
Methods: A total of 38 Postmortem OFC samples were obtained from the VA Brain Bank (OUD = 12; Control = 26). mCpHs were assessed using reduced representation oxidative bisulfite sequencing in neuronal nuclei. Differential analysis was performed using the "methylkit" R package. Age, ancestry, postmortem interval, PTSD, and smoking status were included as covariates. Significant mCpHs were set at q-value < 0.05. Gene Ontology (GO) and KEGG enrichment analyses were performed for the annotated genes of all differential mCpH loci using String, ShinyGO, and amiGO software. Further, all annotated genes were analyzed using the Drug gene interaction database (DGIdb).
Results: A total of 2,352 differentially methylated genome-wide significant mCpHs were identified in OUD, mapping to 2,081 genes. GO analysis of genes with differential mCpH loci showed enrichment for nervous system development (p-value = 2.32E-19). KEGG enrichment analysis identified axon guidance and glutamatergic synapse (FDR 9E-4-2.1E-2). Drug interaction analysis found 3,420 interactions between the annotated genes and drugs, identifying interactions with 15 opioid-related drugs, including lofexidine and tizanidine, both previously used for the treatment of OUD-related symptoms.
Conclusion: Our findings suggest a role of mCpHs for OUD in cortical neurons and reveal important biological pathways and drug targets associated with the disorder.
Keywords: epigenetic; methylation; non-CpG site; opioid; orbitofrontal cortex; postmortem human brain.
Copyright © 2023 Nagamatsu, Rompala, Hurd, Núñez-Rios, Montalvo-Ortiz and Traumatic Stress Brain Research Group.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Deak J, Zhou H, Galimberti M, Levey D, Wendt F, Sanchez-Roige S, et al. Genome-wide association study in individuals of European and African ancestry and multi-trait analysis of opioid use disorder identifies 19 independent genome-wide significant risk loci. Mol Psychiatry. (2022) 27:3970–9. 10.1038/s41380-022-01709-1 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

