Evaluating LC-HRMS metabolomics data processing software using FAIR principles for research software
- PMID: 36745241
- PMCID: PMC12087353
- DOI: 10.1007/s11306-023-01974-3
Evaluating LC-HRMS metabolomics data processing software using FAIR principles for research software
Abstract
Background: Liquid chromatography-high resolution mass spectrometry (LC-HRMS) is a popular approach for metabolomics data acquisition and requires many data processing software tools. The FAIR Principles - Findability, Accessibility, Interoperability, and Reusability - were proposed to promote open science and reusable data management, and to maximize the benefit obtained from contemporary and formal scholarly digital publishing. More recently, the FAIR principles were extended to include Research Software (FAIR4RS).
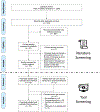
Aim of review: This study facilitates open science in metabolomics by providing an implementation solution for adopting FAIR4RS in the LC-HRMS metabolomics data processing software. We believe our evaluation guidelines and results can help improve the FAIRness of research software.
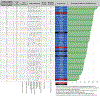
Key scientific concepts of review: We evaluated 124 LC-HRMS metabolomics data processing software obtained from a systematic review and selected 61 software for detailed evaluation using FAIR4RS-related criteria, which were extracted from the literature along with internal discussions. We assigned each criterion one or more FAIR4RS categories through discussion. The minimum, median, and maximum percentages of criteria fulfillment of software were 21.6%, 47.7%, and 71.8%. Statistical analysis revealed no significant improvement in FAIRness over time. We identified four criteria covering multiple FAIR4RS categories but had a low %fulfillment: (1) No software had semantic annotation of key information; (2) only 6.3% of evaluated software were registered to Zenodo and received DOIs; (3) only 14.5% of selected software had official software containerization or virtual machine; (4) only 16.7% of evaluated software had a fully documented functions in code. According to the results, we discussed improvement strategies and future directions.
Keywords: FAIR principles; Liquid chromatography-mass spectrometry; Metabolomics; Open science; Reproducibility; Research software.
© 2023. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
Declarations
Figures
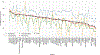


References
-
- Aghamohammadi A, Mirian-Hosseinabadi SH, & Jalali S (2021). Statement frequency coverage: a code coverage criterion for assessing test suite effectiveness. Information and Software Technology, 129, 106426. 10.1016/j.infsof.2020.106426. - DOI
-
- Analytica Chimica Acta | Journal | ScienceDirect.com by Elsevier. (n.d.). Retrieved September 16, from https://www.sciencedirect.com/journal/analytica-chimica-acta
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
