Evaluation of sequencing and PCR-based methods for the quantification of the viral genome formula
- PMID: 36746340
- PMCID: PMC10194290
- DOI: 10.1016/j.virusres.2023.199064
Evaluation of sequencing and PCR-based methods for the quantification of the viral genome formula
Abstract
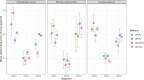
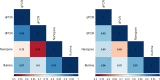
Viruses show great diversity in their genome organization. Multipartite viruses package their genome segments into separate particles, most or all of which are required to initiate infection in the host cell. The benefits of such seemingly inefficient genome organization are not well understood. One hypothesised benefit of multipartition is that it allows for flexible changes in gene expression by altering the frequency of each genome segment in different environments, such as encountering different host species. The ratio of the frequency of segments is termed the genome formula (GF). Thus far, formal studies quantifying the GF have been performed for well-characterised virus-host systems in experimental settings using RT-qPCR. However, to understand GF variation in natural populations or novel virus-host systems, a comparison of several methods for GF estimation including high-throughput sequencing (HTS) based methods is needed. Currently, it is unclear how HTS-methods compare a golden standard, such as RT-qPCR. Here we show a comparison of multiple GF quantification methods (RT-qPCR, RT-digital PCR, Illumina RNAseq and Nanopore direct RNA sequencing) using three host plants (Nicotiana tabacum, Nicotiana benthamiana, and Chenopodium quinoa) infected with cucumber mosaic virus (CMV), a tripartite RNA virus. Our results show that all methods give roughly similar results, though there is a significant method effect on genome formula estimates. While the RT-qPCR and RT-dPCR GF estimates are congruent, the GF estimates from HTS methods deviate from those found with PCR. Our findings emphasize the need to tailor the GF quantification method to the experimental aim, and highlight that it may not be possible to compare HTS and PCR-based methods directly. The difference in results between PCR-based methods and HTS highlights that the choice of quantification technique is not trivial.
Keywords: Cucumber mosaic virus; Digital PCR; Genome formula; Long-read sequencing; Multipartite virus; Quantitative PCR.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Akaike H. A New Look at the Statistical Model Identification. IEEE Trans. Autom. Contr. 1974;19(6):716–723.
-
- Ben Chehida S., Filloux D., Fernandez E., Moubset O., Hoareau M., Julian C., Blondin L., Lett J.M., Roumagnac P., Lefeuvre P. Nanopore sequencing is a credible alternative to recover complete genomes of geminiviruses. Microorganisms. 2021;9(5) doi: 10.3390/microorganisms9050903. - DOI - PMC - PubMed
-
- Bronzato Badial A., Sherman D., Stone A., Gopakumar A., Wilson V., Schneider W., King J. Nanopore sequencing as a surveillance tool for plant pathogens in plant and insect tissues. Plant Dis. 2018;102(8):1648–1652. - PubMed
-
- Bustin SA., Benes V., Garson JA., Hellemans J., Huggett J., Kubista M., Mueller R., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

