LSD1/PRMT6-targeting gene therapy to attenuate androgen receptor toxic gain-of-function ameliorates spinobulbar muscular atrophy phenotypes in flies and mice
- PMID: 36746939
- PMCID: PMC9902531
- DOI: 10.1038/s41467-023-36186-9
LSD1/PRMT6-targeting gene therapy to attenuate androgen receptor toxic gain-of-function ameliorates spinobulbar muscular atrophy phenotypes in flies and mice
Abstract
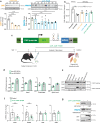
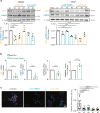
Spinobulbar muscular atrophy (SBMA) is caused by CAG expansions in the androgen receptor gene. Androgen binding to polyQ-expanded androgen receptor triggers SBMA through a combination of toxic gain-of-function and loss-of-function mechanisms. Leveraging cell lines, mice, and patient-derived specimens, we show that androgen receptor co-regulators lysine-specific demethylase 1 (LSD1) and protein arginine methyltransferase 6 (PRMT6) are overexpressed in an androgen-dependent manner specifically in the skeletal muscle of SBMA patients and mice. LSD1 and PRMT6 cooperatively and synergistically transactivate androgen receptor, and their effect is enhanced by expanded polyQ. Pharmacological and genetic silencing of LSD1 and PRMT6 attenuates polyQ-expanded androgen receptor transactivation in SBMA cells and suppresses toxicity in SBMA flies, and a preclinical approach based on miRNA-mediated silencing of LSD1 and PRMT6 attenuates disease manifestations in SBMA mice. These observations suggest that targeting overexpressed co-regulators can attenuate androgen receptor toxic gain-of-function without exacerbating loss-of-function, highlighting a potential therapeutic strategy for patients with SBMA.
© 2023. The Author(s).
Conflict of interest statement
M.P., M.B., G.R., and A.C. are named as co-inventors on the patent application Italian Priority N. 102022000026595 “New inhibitors of epigenetic regulators/nuovi inibitori di regolatori epigenetici”. The other authors declare no competing interests.
Figures








References
-
- Kawahara, H. A family of progressive bulbar palsy. Aichi. Med. J.16, 3–4 (1897).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

