Newly identified sex chromosomes in the Sphagnum (peat moss) genome alter carbon sequestration and ecosystem dynamics
- PMID: 36747050
- PMCID: PMC9946827
- DOI: 10.1038/s41477-022-01333-5
Newly identified sex chromosomes in the Sphagnum (peat moss) genome alter carbon sequestration and ecosystem dynamics
Abstract
Peatlands are crucial sinks for atmospheric carbon but are critically threatened due to warming climates. Sphagnum (peat moss) species are keystone members of peatland communities where they actively engineer hyperacidic conditions, which improves their competitive advantage and accelerates ecosystem-level carbon sequestration. To dissect the molecular and physiological sources of this unique biology, we generated chromosome-scale genomes of two Sphagnum species: S. divinum and S. angustifolium. Sphagnum genomes show no gene colinearity with any other reference genome to date, demonstrating that Sphagnum represents an unsampled lineage of land plant evolution. The genomes also revealed an average recombination rate an order of magnitude higher than vascular land plants and short putative U/V sex chromosomes. These newly described sex chromosomes interact with autosomal loci that significantly impact growth across diverse pH conditions. This discovery demonstrates that the ability of Sphagnum to sequester carbon in acidic peat bogs is mediated by interactions between sex, autosomes and environment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
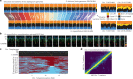



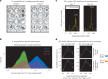
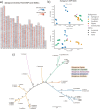


Comment in
-
Sex chromosomes: How to make a hermaphrodite.Curr Biol. 2023 Nov 6;33(21):R1150-R1152. doi: 10.1016/j.cub.2023.09.038. Curr Biol. 2023. PMID: 37935128
References
-
- Yu, Z., Loisel, J., Brosseau, D. P., Beilman, D. W. & Hunt, S. J. Global peatland dynamics since the Last Glacial Maximum. Geophys. Res. Lett.37, L13402 (2010).
-
- Piatkowski, B. T. From Genes to Traits and Ecosystems: Evolutionary Ecology of Sphagnum (Peat Moss). PhD dissertation, Duke Univ. (2020).
-
- Rydin, H. & Jeglum, J. K. The Biology of Peatlands (Oxford Univ. Press, 2013).

