This is a preprint.
Structural variation across 138,134 samples in the TOPMed consortium
- PMID: 36747810
- PMCID: PMC9900832
- DOI: 10.1101/2023.01.25.525428
Structural variation across 138,134 samples in the TOPMed consortium
Abstract
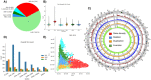
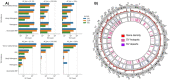
Ever larger Structural Variant (SV) catalogs highlighting the diversity within and between populations help researchers better understand the links between SVs and disease. The identification of SVs from DNA sequence data is non-trivial and requires a balance between comprehensiveness and precision. Here we present a catalog of 355,667 SVs (59.34% novel) across autosomes and the X chromosome (50bp+) from 138,134 individuals in the diverse TOPMed consortium. We describe our methodologies for SV inference resulting in high variant quality and >90% allele concordance compared to long-read de-novo assemblies of well-characterized control samples. We demonstrate utility through significant associations between SVs and important various cardio-metabolic and hemotologic traits. We have identified 690 SV hotspots and deserts and those that potentially impact the regulation of medically relevant genes. This catalog characterizes SVs across multiple populations and will serve as a valuable tool to understand the impact of SV on disease development and progression.
Keywords: NGS; Population; Structural Variants; TOPMed.
Figures


References
Publication types
Grants and funding
- N01 HC095160/HL/NHLBI NIH HHS/United States
- U01 HL054472/HL/NHLBI NIH HHS/United States
- R01 HL113264/HL/NHLBI NIH HHS/United States
- U01 HG007417/HG/NHGRI NIH HHS/United States
- UM1 HL098162/HL/NHLBI NIH HHS/United States
- R01 ES021801/ES/NIEHS NIH HHS/United States
- R01 ES015794/ES/NIEHS NIH HHS/United States
- K99 ES027870/ES/NIEHS NIH HHS/United States
- 75N92020D00002/HL/NHLBI NIH HHS/United States
- HHSN268201300005C/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- R01 HL113323/HL/NHLBI NIH HHS/United States
- UL1 RR024989/RR/NCRR NIH HHS/United States
- 75N92021D00002/HL/NHLBI NIH HHS/United States
- R01 HL095080/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- P30 ES007048/ES/NIEHS NIH HHS/United States
- HHSN268201800012C/HL/NHLBI NIH HHS/United States
- HHSN268201300004C/HL/NHLBI NIH HHS/United States
- N01 HC095161/HL/NHLBI NIH HHS/United States
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- R01 HL104135/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 HL123915/HL/NHLBI NIH HHS/United States
- K23 HL004464/HL/NHLBI NIH HHS/United States
- UM1 HL098123/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R01 ES016535/ES/NIEHS NIH HHS/United States
- R01 HL046380/HL/NHLBI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- R01 HL083141/HL/NHLBI NIH HHS/United States
- 75N92021D00005/WH/WHI NIH HHS/United States
- R01 HL121007/HL/NHLBI NIH HHS/United States
- R01 HL155742/HL/NHLBI NIH HHS/United States
- U01 DK085524/DK/NIDDK NIH HHS/United States
- R01 HL076647/HL/NHLBI NIH HHS/United States
- R01 HL111249/HL/NHLBI NIH HHS/United States
- HHSN268201300001C/HL/NHLBI NIH HHS/United States
- HHSN268201300003I/HL/NHLBI NIH HHS/United States
- U01 HL054527/HL/NHLBI NIH HHS/United States
- HHSN268201800004I/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- HHSN268201800014I/HB/NHLBI NIH HHS/United States
- N01 HC065233/HL/NHLBI NIH HHS/United States
- R01 HL068986/HL/NHLBI NIH HHS/United States
- R01 HL043851/HL/NHLBI NIH HHS/United States
- N01 HC065236/HL/NHLBI NIH HHS/United States
- T32 HL007185/HL/NHLBI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- R01 HL087660/HL/NHLBI NIH HHS/United States
- R01 AR048797/AR/NIAMS NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- U10 HL054464/HL/NHLBI NIH HHS/United States
- N01 HC065235/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- RC2 HL101543/HL/NHLBI NIH HHS/United States
- P01 ES009581/ES/NIEHS NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- R01 HL085251/HL/NHLBI NIH HHS/United States
- R01 HL066216/HL/NHLBI NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- U01 HL054509/HL/NHLBI NIH HHS/United States
- 75N92020D00001/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- P01 ES022845/ES/NIEHS NIH HHS/United States
- R01 HL061768/HL/NHLBI NIH HHS/United States
- R01 HL113338/HL/NHLBI NIH HHS/United States
- P50 CA180905/CA/NCI NIH HHS/United States
- R01 AG058921/AG/NIA NIH HHS/United States
- R01 NS058700/NS/NINDS NIH HHS/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- U01 HL131003/HL/NHLBI NIH HHS/United States
- HHSN268201800014C/HL/NHLBI NIH HHS/United States
- 75N92020D00003/HL/NHLBI NIH HHS/United States
- F32 HL085989/HL/NHLBI NIH HHS/United States
- R01 MH078111/MH/NIMH NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- U01 HL054464/HL/NHLBI NIH HHS/United States
- R01 HL090620/HL/NHLBI NIH HHS/United States
- R01 HL119443/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- 75N92021D00001/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- P30 GM110766/GM/NIGMS NIH HHS/United States
- R01 HL067348/HL/NHLBI NIH HHS/United States
- U10 HL054457/HL/NHLBI NIH HHS/United States
- R35 HL135818/HL/NHLBI NIH HHS/United States
- P30 AG015272/AG/NIA NIH HHS/United States
- HHSN268201800003I/HL/NHLBI NIH HHS/United States
- R01 HL113326/HL/NHLBI NIH HHS/United States
- U10 HL054481/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- RC2 HL101651/HL/NHLBI NIH HHS/United States
- U01 HL072524/HL/NHLBI NIH HHS/United States
- R01 HL146860/HL/NHLBI NIH HHS/United States
- HHSN268201800007I/HL/NHLBI NIH HHS/United States
- N01 HC065234/HL/NHLBI NIH HHS/United States
- HHSN268201700002C/HL/NHLBI NIH HHS/United States
- P01 ES011627/ES/NIEHS NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- R01 HL087699/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201800013I/MD/NIMHD NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- U01 HL054457/HL/NHLBI NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- R01 HL111314/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- R01 HL087680/HL/NHLBI NIH HHS/United States
- HHSN268201800012I/HL/NHLBI NIH HHS/United States
- 75N92021D00003/WH/WHI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- 75N92020D00004/HL/NHLBI NIH HHS/United States
- U01 HL054498/HL/NHLBI NIH HHS/United States
- R01 HL104608/HL/NHLBI NIH HHS/United States
- U01 HL072515/HL/NHLBI NIH HHS/United States
- P01 HL045522/HL/NHLBI NIH HHS/United States
- UM1 CA182913/CA/NCI NIH HHS/United States
- UM1 HL128761/HL/NHLBI NIH HHS/United States
- 75N92020D00007/HL/NHLBI NIH HHS/United States
- KL2 RR024990/RR/NCRR NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- HHSN268201800011C/HL/NHLBI NIH HHS/United States
- HHSN268201300003C/HG/NHGRI NIH HHS/United States
- M01 RR007122/RR/NCRR NIH HHS/United States
- R01 CA047988/CA/NCI NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- R01 ES023262/ES/NIEHS NIH HHS/United States
- 75N92021D00006/HL/NHLBI NIH HHS/United States
- R01 HL087263/HL/NHLBI NIH HHS/United States
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- R01 HL080467/HL/NHLBI NIH HHS/United States
- R01 HL068959/HL/NHLBI NIH HHS/United States
- U01 HL072507/HL/NHLBI NIH HHS/United States
- R03 ES014046/ES/NIEHS NIH HHS/United States
- R01 DK071891/DK/NIDDK NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- R01 MH078143/MH/NIMH NIH HHS/United States
- U01 HG004735/HG/NHGRI NIH HHS/United States
- R01 HL117004/HL/NHLBI NIH HHS/United States
- HHSN268201800015I/HB/NHLBI NIH HHS/United States
- U01 HL054481/HL/NHLBI NIH HHS/United States
- 75N92019D00031/HL/NHLBI NIH HHS/United States
- HHSN268201700004C/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- UM1 HL128711/HL/NHLBI NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- R01 HL151855/HL/NHLBI NIH HHS/United States
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- R01 MH083824/MH/NIMH NIH HHS/United States
- HHSN268201800010I/HB/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- UM1 HL098147/HL/NHLBI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- 75N92020D00006/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- R01 HL073410/HL/NHLBI NIH HHS/United States
- HHSN268201800005I/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States
- R01 HL090682/HL/NHLBI NIH HHS/United States
- HHSN268201800011I/HB/NHLBI NIH HHS/United States
- RC1 HL099355/HL/NHLBI NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- N01 HC065237/HL/NHLBI NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- HHSN268201800006I/HL/NHLBI NIH HHS/United States
- U01 HL054495/HL/NHLBI NIH HHS/United States
- R01 AG018728/AG/NIA NIH HHS/United States
- 75N92021D00004/WH/WHI NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- R01 HL055673/HL/NHLBI NIH HHS/United States
- R01 HL079915/HL/NHLBI NIH HHS/United States
- R01 HL092301/HL/NHLBI NIH HHS/United States
- U01 HL054473/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
