This is a preprint.
Reprogramming the Human Gut Microbiome Reduces Dietary Energy Harvest
- PMID: 36747835
- PMCID: PMC9901041
- DOI: 10.21203/rs.3.rs-2382790/v1
Reprogramming the Human Gut Microbiome Reduces Dietary Energy Harvest
Update in
-
Host-diet-gut microbiome interactions influence human energy balance: a randomized clinical trial.Nat Commun. 2023 May 31;14(1):3161. doi: 10.1038/s41467-023-38778-x. Nat Commun. 2023. PMID: 37258525 Free PMC article. Clinical Trial.
Abstract
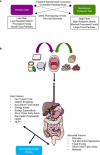
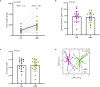
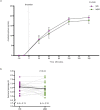
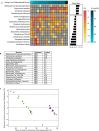
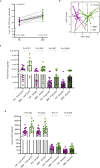
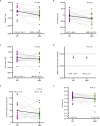
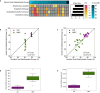
The gut microbiome is emerging as a key modulator of host energy balance1. We conducted a quantitative bioenergetics study aimed at understanding microbial and host factors contributing to energy balance. We used a Microbiome Enhancer Diet (MBD) to reprogram the gut microbiome by delivering more dietary substrates to the colon and randomized healthy participants into a within-subject crossover study with a Western Diet (WD) as a comparator. In a metabolic ward where the environment was strictly controlled, we measured energy intake, energy expenditure, and energy output (fecal, urinary, and methane)2. The primary endpoint was the within-participant difference in host metabolizable energy between experimental conditions. The MBD led to an additional 116 ± 56 kcals lost in feces daily and thus, lower metabolizable energy for the host by channeling more energy to the colon and microbes. The MBD drove significant shifts in microbial biomass, community structure, and fermentation, with parallel alterations to the host enteroendocrine system and without altering appetite or energy expenditure. Host metabolizable energy on the MBD had quantitatively significant interindividual variability, which was associated with differences in the composition of the gut microbiota experimentally and colonic transit time and short-chain fatty acid absorption in silico. Our results provide key insights into how a diet designed to optimize the gut microbiome lowers host metabolizable energy in healthy humans.
Conflict of interest statement
Figures











References
Methods References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

