A Monoclonal Human Alveolar Epithelial Cell Line ("Arlo") with Pronounced Barrier Function for Studying Drug Permeability and Viral Infections
- PMID: 36748276
- PMCID: PMC10015904
- DOI: 10.1002/advs.202207301
A Monoclonal Human Alveolar Epithelial Cell Line ("Arlo") with Pronounced Barrier Function for Studying Drug Permeability and Viral Infections
Abstract
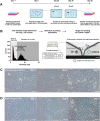
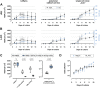
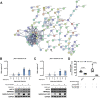
In the development of orally inhaled drug products preclinical animal models regularly fail to predict pharmacological as well as toxicological responses in humans. Models based on human cells and tissues are potential alternatives to animal experimentation allowing for the isolation of essential processes of human biology and making them accessible in vitro. Here, the generation of a novel monoclonal cell line "Arlo," derived from the polyclonal human alveolar epithelium lentivirus immortalized cell line hAELVi via single-cell printing, and its characterization as a model for the human alveolar epithelium as well as a building block for future complex in vitro models is described. "Arlo" is systematically compared in vitro to primary human alveolar epithelial cells (hAEpCs) as well as to the polyclonal hAELVi cell line. "Arlo" cells show enhanced barrier properties with high transepithelial electrical resistance (TEER) of ≈3000 Ω cm2 and a potential difference (PD) of ≈30 mV under air-liquid interface (ALI) conditions, that can be modulated. The cells grow in a polarized monolayer and express genes relevant to barrier integrity as well as homeostasis as is observed in hAEpCs. Successful productive infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a proof-of-principle study offers an additional, attractive application of "Arlo" beyond biopharmaceutical experimentation.
Keywords: Transwell inserts; drug transport; lung; pulmonary drug delivery; tight junctions.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
C.‐M.L., N.S.‐D., and P.C. are the creators of the cell line “Arlo”. A manufacture and distribution license for the cell line “Arlo” was granted to InSCREENeX GmbH, Germany by the Helmholtz Centre for Infection Research (Helmholtz‐Zentrum für Infektionsforschung GmbH) (HZI), Germany. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Seok J., Warren H. S., Cuenca A. G., Mindrinos M. N., Baker H. V., Xu W., Richards D. R., McDonald‐Smith G. P., Gao H., Hennessy L., Finnerty C. C., López C. M., Honari S., Moore E. E., Minei J. P., Cuschieri J., Bankey P. E., Johnson J. L., Sperry J., Nathens A. B., Billiar T. R., West M. A., Jeschke M. G., Klein M. B., Gamelli R. L., Gibran N. S., Brownstein B. H., Miller‐Graziano C., Calvano S. E., Mason P. H., et al., Proc. Natl. Acad. Sci. USA 2013, 110, 3507. - PMC - PubMed
-
- Carius P., Horstmann J. C., Carvalho‐Wodarz S., de C., Lehr C.‐M., Handb. Exp. Pharmacol. 2021, 265, 157. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
