Genome-wide association studies reveal distinct genetic correlates and increased heritability of antimicrobial resistance in Vibrio cholerae under anaerobic conditions
- PMID: 36748512
- PMCID: PMC9837564
- DOI: 10.1099/mgen.0.000905
Genome-wide association studies reveal distinct genetic correlates and increased heritability of antimicrobial resistance in Vibrio cholerae under anaerobic conditions
Abstract
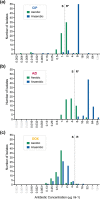
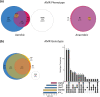
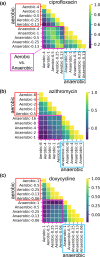
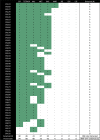
The antibiotic formulary is threatened by high rates of antimicrobial resistance (AMR) among enteropathogens. Enteric bacteria are exposed to anaerobic conditions within the gastrointestinal tract, yet little is known about how oxygen exposure influences AMR. The facultative anaerobe Vibrio cholerae was chosen as a model to address this knowledge gap. We obtained V. cholerae isolates from 66 cholera patients, sequenced their genomes, and grew them under anaerobic and aerobic conditions with and without three clinically relevant antibiotics (ciprofloxacin, azithromycin, doxycycline). For ciprofloxacin and azithromycin, the minimum inhibitory concentration (MIC) increased under anaerobic conditions compared to aerobic conditions. Using standard resistance breakpoints, the odds of classifying isolates as resistant increased over 10 times for ciprofloxacin and 100 times for azithromycin under anaerobic conditions compared to aerobic conditions. For doxycycline, nearly all isolates were sensitive under both conditions. Using genome-wide association studies, we found associations between genetic elements and AMR phenotypes that varied by oxygen exposure and antibiotic concentrations. These AMR phenotypes were more heritable, and the AMR-associated genetic elements were more often discovered, under anaerobic conditions. These AMR-associated genetic elements are promising targets for future mechanistic research. Our findings provide a rationale to determine whether increased MICs under anaerobic conditions are associated with therapeutic failures and/or microbial escape in cholera patients. If so, there may be a need to determine new AMR breakpoints for anaerobic conditions.
Keywords: Vibrio cholerae; anaerobic; antibiotics; antimicrobial resistance; cholera; enteropathogens.
Conflict of interest statement
The authors declare that there are no conflicts of interest
Figures

References
-
- CLSI Performance Standards for Antimicrobial Susceptibility Testing, 27th edn. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

