The DataHarmonizer: a tool for faster data harmonization, validation, aggregation and analysis of pathogen genomics contextual information
- PMID: 36748616
- PMCID: PMC9973856
- DOI: 10.1099/mgen.0.000908
The DataHarmonizer: a tool for faster data harmonization, validation, aggregation and analysis of pathogen genomics contextual information
Abstract
Pathogen genomics is a critical tool for public health surveillance, infection control, outbreak investigations as well as research. In order to make use of pathogen genomics data, they must be interpreted using contextual data (metadata). Contextual data include sample metadata, laboratory methods, patient demographics, clinical outcomes and epidemiological information. However, the variability in how contextual information is captured by different authorities and how it is encoded in different databases poses challenges for data interpretation, integration and their use/re-use. The DataHarmonizer is a template-driven spreadsheet application for harmonizing, validating and transforming genomics contextual data into submission-ready formats for public or private repositories. The tool's web browser-based JavaScript environment enables validation and its offline functionality and local installation increases data security. The DataHarmonizer was developed to address the data sharing needs that arose during the COVID-19 pandemic, and was used by members of the Canadian COVID Genomics Network (CanCOGeN) to harmonize SARS-CoV-2 contextual data for national surveillance and for public repository submission. In order to support coordination of international surveillance efforts, we have partnered with the Public Health Alliance for Genomic Epidemiology to also provide a template conforming to its SARS-CoV-2 contextual data specification for use worldwide. Templates are also being developed for One Health and foodborne pathogens. Overall, the DataHarmonizer tool improves the effectiveness and fidelity of contextual data capture as well as its subsequent usability. Harmonization of contextual information across authorities, platforms and systems globally improves interoperability and reusability of data for concerted public health and research initiatives to fight the current pandemic and future public health emergencies. While initially developed for the COVID-19 pandemic, its expansion to other data management applications and pathogens is already underway.
Keywords: contextual data; data management; genomic surveillance; harmonization; metadata.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
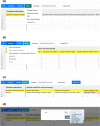


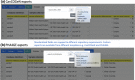
References
-
- Zhang W, Govindavari JP, Davis BD, Chen SS, Kim JT, et al. Analysis of genomic characteristics and transmission routes of patients with confirmed SARS-CoV-2 in Southern California during the early stage of the US COVID-19 pandemic. JAMA Netw Open. 2020;3:e2024191. doi: 10.1001/jamanetworkopen.2020.24191. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

