SLAMF7 regulates the inflammatory response in macrophages during polymicrobial sepsis
- PMID: 36749634
- PMCID: PMC10014109
- DOI: 10.1172/JCI150224
SLAMF7 regulates the inflammatory response in macrophages during polymicrobial sepsis
Abstract
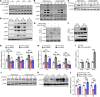
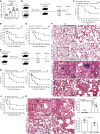
Uncontrolled inflammation occurred in sepsis results in multiple organ injuries and shock, which contributes to the death of patients with sepsis. However, the regulatory mechanisms that restrict excessive inflammation are still elusive. Here, we identified an Ig-like receptor called signaling lymphocyte activation molecular family 7 (SLAMF7) as a key suppressor of inflammation during sepsis. We found that the expression of SLAMF7 on monocytes/macrophages was significantly elevated in patients with sepsis and in septic mice. SLAMF7 attenuated TLR-dependent MAPK and NF-κB signaling activation in macrophages by cooperating with Src homology 2-containing inositol-5'‑phosphatase 1 (SHIP1). Furthermore, SLAMF7 interacted with SHIP1 and TNF receptor-associated factor 6 (TRAF6) to inhibit K63 ubiquitination of TRAF6. In addition, we found that tyrosine phosphorylation sites within the intracellular domain of SLAMF7 and the phosphatase domain of SHIP1 were indispensable for the interaction between SLAMF7, SHIP1, and TRAF6 and SLAMF7-mediated modulation of cytokine production. Finally, we demonstrated that SLAMF7 protected against lethal sepsis and endotoxemia by downregulating macrophage proinflammatory cytokines and suppressing inflammation-induced organ damage. Taken together, our findings reveal a negative regulatory role of SLAMF7 in polymicrobial sepsis, thus providing sights into the treatment of sepsis.
Keywords: Bacterial infections; Infectious disease; Inflammation; Macrophages.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

