Analysis of 363 Genetic Variants in F5 via an Interactive Web Database Reveals New Insights into FV Deficiency and FV Leiden
- PMID: 36751301
- PMCID: PMC9829979
- DOI: 10.1055/a-1987-5978
Analysis of 363 Genetic Variants in F5 via an Interactive Web Database Reveals New Insights into FV Deficiency and FV Leiden
Abstract
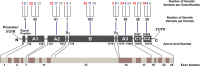
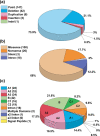
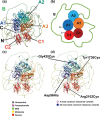
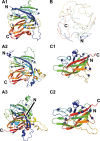
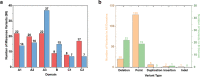
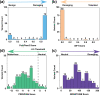
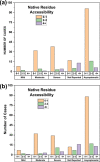
The inherited bleeding disorder Factor V (FV) deficiency and clotting risk factor FV Leiden are associated with genetic variants in the F5 gene. FV deficiency occurs with mild, moderate, severe, or asymptomatic phenotypes, and either dysfunctional or reduced amounts of plasma FV protein. Here we present an interactive web database containing 363 unique F5 variants derived from 801 patient records, with 199 FV deficiency-associated variants from 245 patient records. Their occurrence is rationalized based on the 2,224 residue sequence and new FV protein structures. The 199 FV deficiency variants correspond to 26 (13%) mild, 22 (11%) moderate, 49 (25%) severe, 35 (18%) asymptomatic, and 67 (34%) unreported phenotypes. Their variant distributions in the FV domains A1, A2, A3, B, C1 and C2 were 28 (14%), 32 (16%), 34 (17%), 42 (21%), 16 (8%), and 19 variants (10%), respectively, showing that these six regions contain similar proportions of variants. Variants associated with FV deficiency do not cluster near known protein-partner binding sites, thus the molecular mechanism leading to the phenotypes cannot be explained. However, the widespread distribution of FV variants in combination with a high proportion of buried variant residues indicated that FV is susceptible to disruption by small perturbations in its globular structure. Variants located in the disordered B domain also appear to disrupt the FV structure. We discuss how the interactive database provides an online resource that clarifies the clinical understanding of FV deficiency.
Keywords: coagulation; coagulation factors; gene mutations; protein structure.
The Author(s). This is an open access article published by Thieme under the terms of the Creative Commons Attribution License, permitting unrestricted use, distribution, and reproduction so long as the original work is properly cited. ( https://creativecommons.org/licenses/by/4.0/ ).
Conflict of interest statement
Conflict of Interest None declared.
Figures
Similar articles
-
Next generation sequencing in bleeding disorders: two novel variants in the F5 gene (Valencia-1 and Valencia-2) associated with mild factor V deficiency.J Thromb Thrombolysis. 2019 Nov;48(4):674-678. doi: 10.1007/s11239-019-01911-z. J Thromb Thrombolysis. 2019. PMID: 31267299
-
Significance of the p.Phe218Ser and p.Gly304Glu F5 Variants in Hereditary Factor V Deficiency.Acta Haematol. 2021;144(6):712-716. doi: 10.1159/000512363. Epub 2021 Jul 19. Acta Haematol. 2021. PMID: 34280927
-
Analysis of 180 Genetic Variants in a New Interactive FX Variant Database Reveals Novel Insights into FX Deficiency.TH Open. 2021 Nov 23;5(4):e557-e569. doi: 10.1055/a-1704-0841. eCollection 2021 Oct. TH Open. 2021. PMID: 35059555 Free PMC article.
-
Pro- and anticoagulant properties of factor V in pathogenesis of thrombosis and bleeding disorders.Int J Lab Hematol. 2016 May;38 Suppl 1:4-11. doi: 10.1111/ijlh.12508. Epub 2016 May 9. Int J Lab Hematol. 2016. PMID: 27161771 Review.
-
Factor V deficiency.Semin Thromb Hemost. 2009 Jun;35(4):382-9. doi: 10.1055/s-0029-1225760. Epub 2009 Jul 13. Semin Thromb Hemost. 2009. PMID: 19598066 Review.
Cited by
-
[Pedigree analysis of novel missense mutations causing hereditary coagulation factor Ⅴ deficiency].Zhonghua Xue Ye Xue Za Zhi. 2024 Dec 14;45(12):1119-1124. doi: 10.3760/cma.j.cn121090-20240424-00161-1. Zhonghua Xue Ye Xue Za Zhi. 2024. PMID: 39765353 Free PMC article. Chinese.
-
Factor V variants in bleeding and thrombosis.Res Pract Thromb Haemost. 2024 Jan 26;8(1):102330. doi: 10.1016/j.rpth.2024.102330. eCollection 2024 Jan. Res Pract Thromb Haemost. 2024. PMID: 38404937 Free PMC article.
-
F5 6665A>G Polymorphism Is Associated with Increased Risk of Venous Thromboembolism in Females.Int J Mol Sci. 2025 Mar 7;26(6):2403. doi: 10.3390/ijms26062403. Int J Mol Sci. 2025. PMID: 40141044 Free PMC article.
-
Analyzing 6211 unique variants in the upgraded interactive FVIII web database reveals novel insights into hemophilia A.Blood Vessel Thromb Hemost. 2025 Jan 21;2(3):100053. doi: 10.1016/j.bvth.2025.100053. eCollection 2025 Aug. Blood Vessel Thromb Hemost. 2025. PMID: 40765911 Free PMC article.
References
-
- Lippi G, Favaloro E J, Montagnana M, Manzato F, Guidi G C, Franchini M. Inherited and acquired factor V deficiency. Blood Coagul Fibrinolysis. 2011;22(03):160–166. - PubMed
-
- Kujovich J L. Factor V Leiden thrombophilia. Genet Med. 2011;13(01):1–16. - PubMed
-
- Duga S, Asselta R, Tenchini M L. Coagulation factor V. Int J Biochem Cell Biol. 2004;36(08):1393–1399. - PubMed
-
- Cripe L D, Moore K D, Kane W H. Structure of the gene for human coagulation factor V. Biochemistry. 1992;31(15):3777–3785. - PubMed
-
- Huang J N, Koerper M A. Factor V deficiency: a concise review. Haemophilia. 2008;14(06):1164–1169. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

