Evolutionary rate of SARS-CoV-2 increases during zoonotic infection of farmed mink
- PMID: 36751428
- PMCID: PMC9896948
- DOI: 10.1093/ve/vead002
Evolutionary rate of SARS-CoV-2 increases during zoonotic infection of farmed mink
Abstract
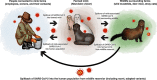
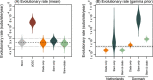
To investigate genetic signatures of adaptation to the mink host, we characterised the evolutionary rate heterogeneity in mink-associated severe acute respiratory syndrome coronaviruses (SARS-CoV-2). In 2020, the first detected anthropozoonotic spillover event of SARS-CoV-2 occurred in mink farms throughout Europe and North America. Both spill-back of mink-associated lineages into the human population and the spread into the surrounding wildlife were reported, highlighting the potential formation of a zoonotic reservoir. Our findings suggest that the evolutionary rate of SARS-CoV-2 underwent an episodic increase upon introduction into the mink host before returning to the normal range observed in humans. Furthermore, SARS-CoV-2 lineages could have circulated in the mink population for a month before detection, and during this period, evolutionary rate estimates were between 3 × 10-3 and 1.05 × 10-2 (95 per cent HPD, with a mean rate of 6.59 × 10-3) a four- to thirteen-fold increase compared to that in humans. As there is evidence for unique mutational patterns within mink-associated lineages, we explored the emergence of four mink-specific Spike protein amino acid substitutions Y453F, S1147L, F486L, and Q314K. We found that mutation Y453F emerged early in multiple mink outbreaks and that mutations F486L and Q314K may co-occur. We suggest that SARS-CoV-2 undergoes a brief, but considerable, increase in evolutionary rate in response to greater selective pressures during species jumps, which may lead to the occurrence of mink-specific mutations. These findings emphasise the necessity of ongoing surveillance of zoonotic SARS-CoV-2 infections in the future.
Keywords: SARS-CoV-2; evolutionary rate; mink; molecular clock; spike gene; spillover.
© The Author(s) 2023. Published by Oxford University Press.
Figures


References
-
- Andrews N. et al. (2021) ‘Effectiveness of COVID-19 Vaccines against the Omicron (B. 1.1. 529) Variant of Concern’, MedRxiv.
-
- Bazykin G. et al. (2021) ‘Emergence of Y453F and Δ69-70HV Mutations in a Lymphoma Patient with Long-Term COVID-19’, Virological.org.
-
- Boni M. F. et al. (2020) ‘Evolutionary Origins of the SARS-CoV-2 Sarbecovirus Lineage Responsible for the COVID-19 Pandemic’, Nature Microbiology, 5: 1408–17. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

