Mobile SARS‑CoV‑2 screening facilities for rapid deployment and university-based diagnostic laboratory
- PMID: 36751470
- PMCID: PMC9893752
- DOI: 10.1002/elsc.202200026
Mobile SARS‑CoV‑2 screening facilities for rapid deployment and university-based diagnostic laboratory
Abstract
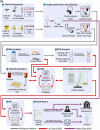
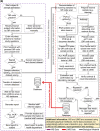
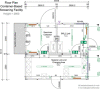
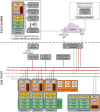
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has created a public crisis. Many medical and public institutions and businesses went into isolation in response to the pandemic. Because SARS-CoV-2 can spread irrespective of a patient's course of disease, these institutions' continued operation or reopening based on the assessment and control of virus spread can be supported by targeted population screening. For this purpose, virus testing in the form of polymerase chain reaction (PCR) analysis and antibody detection in blood can be central. Mobile SARS-CoV-2 screening facilities with a built-in biosafety level (BSL)-2 laboratory were set up to allow the testing offer to be brought close to the subject group's workplace. University staff members, their expertise, and already available equipment were used to implement and operate the screening facilities and a certified diagnostic laboratory. This operation also included specimen collection, transport, PCR and antibody analysis, and informing subjects as well as public health departments. Screening facilities were established at different locations such as educational institutions, nursing homes, and companies providing critical supply chains for health care. Less than 4 weeks after the first imposed lockdown in Germany, a first mobile testing station was established featuring a build-in laboratory with two similar stations commencing operation until June 2020. During the 15-month project period, approximately 33,000 PCR tests and close to 7000 antibody detection tests were collected and analyzed. The presented approach describes the required procedures that enabled the screening facilities and laboratories to collect and process several hundred specimens each day under difficult conditions. This report can assist others in establishing similar setups for pandemic scenarios.
Keywords: COVID‐19; HIS & LIMS; PCR; SARS‐CoV‐2; screening.
© 2023 The Authors. Engineering in Life Sciences published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors have declared no conflicts of interest.
Figures

Similar articles
-
Modelling of hypothetical SARS-CoV-2 point of care tests for routine testing in residential care homes: rapid cost-effectiveness analysis.Health Technol Assess. 2021 Jun;25(39):1-74. doi: 10.3310/hta25390. Health Technol Assess. 2021. PMID: 34142943 Free PMC article.
-
Combined Prospective Seroconversion and PCR Data of Selected Cohorts Indicate a High Rate of Subclinical SARS-CoV-2 Infections-an Open Observational Study in Lower Saxony, Germany.Microbiol Spectr. 2022 Feb 23;10(1):e0151221. doi: 10.1128/spectrum.01512-21. Epub 2022 Feb 16. Microbiol Spectr. 2022. PMID: 35171028 Free PMC article.
-
Safety and Efficacy of Imatinib for Hospitalized Adults with COVID-19: A structured summary of a study protocol for a randomised controlled trial.Trials. 2020 Oct 28;21(1):897. doi: 10.1186/s13063-020-04819-9. Trials. 2020. PMID: 33115543 Free PMC article.
-
Universal screening for SARS-CoV-2 infection: a rapid review.Cochrane Database Syst Rev. 2020 Sep 15;9(9):CD013718. doi: 10.1002/14651858.CD013718. Cochrane Database Syst Rev. 2020. PMID: 33502003 Free PMC article.
-
Intensive diagnostic management of coronavirus disease 2019 (COVID-19) in academic settings in Japan: challenge and future.Inflamm Regen. 2020 Oct 12;40:38. doi: 10.1186/s41232-020-00147-2. eCollection 2020. Inflamm Regen. 2020. PMID: 33062076 Free PMC article. Review.
Cited by
-
A systematic literature review on public health and healthcare resources for pandemic preparedness planning.BMC Public Health. 2024 Nov 11;24(1):3114. doi: 10.1186/s12889-024-20629-z. BMC Public Health. 2024. PMID: 39529010 Free PMC article.
-
Low impact of regular PCR testing on presence at work site during the COVID-19 pandemic: experiences during an open observational study in Lower Saxony 2020-21.BMC Public Health. 2023 Feb 3;23(1):240. doi: 10.1186/s12889-023-15036-9. BMC Public Health. 2023. PMID: 36737718 Free PMC article.
-
Existing operational standards for field deployments of rapid response mobile laboratories: a scoping review.Front Public Health. 2024 Nov 13;12:1455738. doi: 10.3389/fpubh.2024.1455738. eCollection 2024. Front Public Health. 2024. PMID: 39606067 Free PMC article.
References
-
- Stanislawski N, Lange F, Fahnemann C, et al. MCA Dataset, LUIS, 2022. doi:10.25835/jt0dobsz - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous
