ARAX: a graph-based modular reasoning tool for translational biomedicine
- PMID: 36752514
- PMCID: PMC10027432
- DOI: 10.1093/bioinformatics/btad082
ARAX: a graph-based modular reasoning tool for translational biomedicine
Abstract
Motivation: With the rapidly growing volume of knowledge and data in biomedical databases, improved methods for knowledge-graph-based computational reasoning are needed in order to answer translational questions. Previous efforts to solve such challenging computational reasoning problems have contributed tools and approaches, but progress has been hindered by the lack of an expressive analysis workflow language for translational reasoning and by the lack of a reasoning engine-supporting that language-that federates semantically integrated knowledge-bases.
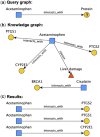
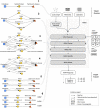
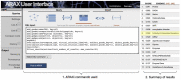
Results: We introduce ARAX, a new reasoning system for translational biomedicine that provides a web browser user interface and an application programming interface (API). ARAX enables users to encode translational biomedical questions and to integrate knowledge across sources to answer the user's query and facilitate exploration of results. For ARAX, we developed new approaches to query planning, knowledge-gathering, reasoning and result ranking and dynamically integrate knowledge providers for answering biomedical questions. To illustrate ARAX's application and utility in specific disease contexts, we present several use-case examples.
Availability and implementation: The source code and technical documentation for building the ARAX server-side software and its built-in knowledge database are freely available online (https://github.com/RTXteam/RTX). We provide a hosted ARAX service with a web browser interface at arax.rtx.ai and a web API endpoint at arax.rtx.ai/api/arax/v1.3/ui/.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2023. Published by Oxford University Press.
Figures


References
-
- Angles R., Gutierrez C. (2008) The Expressive Power of SPARQL. In: The Semantic Web - ISWC 2008. Springer, pp. 114–129.
-
- Brown S.H. et al. (2004) VA national drug file reference terminology: A cross-institutional content coverage study. Stud. Health Technol. Inform., 107, 477–481. - PubMed

