Identification of genes influencing the evolution of Escherichia coli ST372 in dogs and humans
- PMID: 36752777
- PMCID: PMC9997745
- DOI: 10.1099/mgen.0.000930
Identification of genes influencing the evolution of Escherichia coli ST372 in dogs and humans
Erratum in
-
Corrigendum: Identification of genes influencing the evolution of Escherichia coli ST372 in dogs and humans.Microb Genom. 2024 Aug;10(8):001286. doi: 10.1099/mgen.0.001286. Microb Genom. 2024. PMID: 39166982 Free PMC article. No abstract available.
Abstract
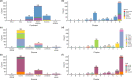
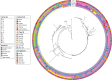
ST372 are widely reported as the major Escherichia coli sequence type in dogs globally. They are also a sporadic cause of extraintestinal infections in humans. Despite this, it is unknown whether ST372 strains from dogs and humans represent shared or distinct populations. Furthermore, little is known about genomic traits that might explain the prominence of ST372 in dogs or presence in humans. To address this, we applied a variety of bioinformatics analyses to a global collection of 407 ST372 E. coli whole-genome sequences to characterize their epidemiological features, population structure and associated accessory genomes. We confirm that dogs are the dominant host of ST372 and that clusters within the population structure exhibit distinctive O:H types. One phylogenetic cluster, 'cluster M', comprised almost half of the sequences and showed the divergence of two human-restricted clades that carried different O:H types to the remainder of the cluster. We also present evidence supporting transmission between dogs and humans within different clusters of the phylogeny, including M. We show that multiple acquisitions of the pdu propanediol utilization operon have occurred in clusters dominated by isolates of canine source, possibly linked to diet, whereas loss of the pdu operon and acquisition of K antigen virulence genes characterize human-restricted lineages.
Keywords: E. coli; ExPEC; ST372; canine; genomic epidemiology; pathogen evolution; pdu operon.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Genomic and Temporal Trends in Canine ExPEC Reflect Those of Human ExPEC.Microbiol Spectr. 2022 Jun 29;10(3):e0129122. doi: 10.1128/spectrum.01291-22. Epub 2022 Jun 8. Microbiol Spectr. 2022. PMID: 35674442 Free PMC article.
-
Genomic analysis of phylogenetic group B2 extraintestinal pathogenic E. coli causing infections in dogs in Australia.Vet Microbiol. 2020 Sep;248:108783. doi: 10.1016/j.vetmic.2020.108783. Epub 2020 Jul 1. Vet Microbiol. 2020. PMID: 32827920
-
Molecular Characteristics of Extraintestinal Pathogenic E. coli (ExPEC), Uropathogenic E. coli (UPEC), and Multidrug Resistant E. coli Isolated from Healthy Dogs in Spain. Whole Genome Sequencing of Canine ST372 Isolates and Comparison with Human Isolates Causing Extraintestinal Infections.Microorganisms. 2020 Oct 31;8(11):1712. doi: 10.3390/microorganisms8111712. Microorganisms. 2020. PMID: 33142871 Free PMC article.
-
Population Structure and Antimicrobial Resistance of Canine Uropathogenic Escherichia coli.J Clin Microbiol. 2018 Aug 27;56(9):e00788-18. doi: 10.1128/JCM.00788-18. Print 2018 Sep. J Clin Microbiol. 2018. PMID: 29997200 Free PMC article.
-
What defines extraintestinal pathogenic Escherichia coli?Int J Med Microbiol. 2011 Dec;301(8):642-7. doi: 10.1016/j.ijmm.2011.09.006. Epub 2011 Oct 5. Int J Med Microbiol. 2011. PMID: 21982038 Review.
Cited by
-
Associations between Isolation Source, Clonal Composition, and Antibiotic Resistance Genes in Escherichia coli Collected in Washington State, USA.Antibiotics (Basel). 2024 Jan 20;13(1):103. doi: 10.3390/antibiotics13010103. Antibiotics (Basel). 2024. PMID: 38275332 Free PMC article.
-
Evolutionary genomic analyses of canine E. coli infections identify a relic capsular locus associated with resistance to multiple classes of antimicrobials.Appl Environ Microbiol. 2024 Aug 21;90(8):e0035424. doi: 10.1128/aem.00354-24. Epub 2024 Jul 16. Appl Environ Microbiol. 2024. PMID: 39012166 Free PMC article.
-
Genotypic Characterization of Uropathogenic Escherichia coli from Companion Animals: Predominance of ST372 in Dogs and Human-Related ST73 in Cats.Antibiotics (Basel). 2023 Dec 30;13(1):38. doi: 10.3390/antibiotics13010038. Antibiotics (Basel). 2023. PMID: 38247597 Free PMC article.
-
Companions in antimicrobial resistance: examining transmission of common antimicrobial-resistant organisms between people and their dogs, cats, and horses.Clin Microbiol Rev. 2025 Mar 13;38(1):e0014622. doi: 10.1128/cmr.00146-22. Epub 2025 Jan 24. Clin Microbiol Rev. 2025. PMID: 39853095 Review.
-
Parameters for one health genomic surveillance of Escherichia coli from Australia.Nat Commun. 2025 Jan 2;16(1):17. doi: 10.1038/s41467-024-55103-2. Nat Commun. 2025. PMID: 39747833 Free PMC article.
References
-
- Bogema DR, McKinnon J, Liu M, Hitchick N, Miller N, et al. Whole-genome analysis of extraintestinal Escherichia coli sequence type 73 from a single hospital over a 2 year period identified different circulating clonal groups. Microb Genom. 2020;6:e000255. doi: 10.1099/mgen.0.000255. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

