Remodeling of colon plasma cell repertoire within ulcerative colitis patients
- PMID: 36752797
- PMCID: PMC9949229
- DOI: 10.1084/jem.20220538
Remodeling of colon plasma cell repertoire within ulcerative colitis patients
Abstract
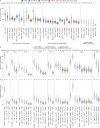
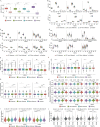
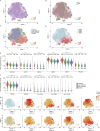
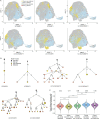
Plasma cells (PCs) constitute a significant fraction of colonic mucosal cells and contribute to inflammatory infiltrates in ulcerative colitis (UC). While gut PCs secrete bacteria-targeting IgA antibodies, their role in UC pathogenesis is unknown. We performed single-cell V(D)J- and RNA-seq on sorted B cells from the colon of healthy individuals and patients with UC. A large fraction of B cell clones is shared between different colon regions, but inflammation in UC broadly disrupts this landscape, causing transcriptomic changes characterized by an increase in the unfolded protein response (UPR) and antigen presentation genes, clonal expansion, and isotype skewing from IgA1 and IgA2 to IgG1. We also directly expressed and assessed the specificity of 152 mAbs from expanded PC clones. These mAbs show low polyreactivity and autoreactivity and instead target both shared bacterial antigens and specific bacterial strains. Altogether, our results characterize the microbiome-specific colon PC response and how its disruption might contribute to inflammation in UC.
© 2023 Scheid et al.
Conflict of interest statement
Disclosures: A.N. Ananthakrishnan reported personal fees from Menten AI and Iterative Scopes outside the submitted work. A. Regev reported “other” from Genentech, Roche, Immunitas, and Celsius Therapeutics; and personal fees from ThermoFisher Scientific and Syros outside the submitted work. In addition, A. Regev had various patents related to single cell genomics issued. R.J. Xavier reported non-financial support from Jnana Therapeutics, Celsius Therapeutics, and MoonLake Immunotherapeutics outside the submitted work. No other disclosures were reported.
Figures








References
-
- Bessissow, T., Lemmens B., Ferrante M., Bisschops R., Van Steen K., Geboes K., Van Assche G., Vermeire S., Rutgeerts P., and De Hertogh G.. 2012. Prognostic value of serologic and histologic markers on clinical relapse in ulcerative colitis patients with mucosal healing. Am. J. Gastroenterol. 107:1684–1692. 10.1038/ajg.2012.301 - DOI - PubMed
-
- Buckner, C.M., Moir S., Kardava L., Ho J., Santich B.H., Kim L.J.Y., Funk E.K., Nelson A.K., Winckler B., Chairez C.L., et al. . 2014. CXCR4/IgG-expressing plasma cells are associated with human gastrointestinal tissue inflammation. J. Allergy Clin. Immunol. 133:1676–1685.e5. 10.1016/j.jaci.2013.10.050 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

