Functional validation of transposable element-derived cis-regulatory elements in Atlantic salmon
- PMID: 36753570
- PMCID: PMC10085797
- DOI: 10.1093/g3journal/jkad034
Functional validation of transposable element-derived cis-regulatory elements in Atlantic salmon
Abstract
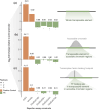
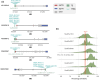
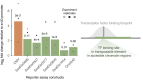
Transposable elements (TEs) are hypothesized to play important roles in shaping genome evolution following whole-genome duplications (WGDs), including rewiring of gene regulation. In a recent analysis, duplicate gene copies that had evolved higher expression in liver following the salmonid WGD ∼100 million years ago were associated with higher numbers of predicted TE-derived cis-regulatory elements (TE-CREs). Yet, the ability of these TE-CREs to recruit transcription factors (TFs) in vivo and impact gene expression remains unknown. Here, we evaluated the gene-regulatory functions of 11 TEs using luciferase promoter reporter assays in Atlantic salmon (Salmo salar) primary liver cells. Canonical Tc1-Mariner elements from intronic regions showed no or small repressive effects on transcription. However, other TE-CREs upstream of transcriptional start sites increased expression significantly. Our results question the hypothesis that TEs in the Tc1-Mariner superfamily, which were extremely active following WGD in salmonids, had a major impact on regulatory rewiring of gene duplicates, but highlights the potential of other TEs in post-WGD rewiring of gene regulation in the Atlantic salmon genome.
Keywords: Atlantic salmon; evolution; gene expression; transposable elements; whole-genome duplication.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Genetics Society of America.
Conflict of interest statement
Conflicts of interest None declared.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
