The catalytic-dead Pcif1 regulates gene expression and fertility in Drosophila
- PMID: 36754578
- PMCID: PMC10158991
- DOI: 10.1261/rna.079192.122
The catalytic-dead Pcif1 regulates gene expression and fertility in Drosophila
Abstract
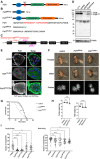
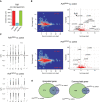
Eukaryotic mRNAs are modified at the 5' end with a methylated guanosine (m7G) that is attached to the transcription start site (TSS) nucleotide. The TSS nucleotide is 2'-O-methylated (Nm) by CMTR1 in organisms ranging from insects to human. In mammals, the TSS adenosine can be further N 6 -methylated by RNA polymerase II phosphorylated CTD-interacting factor 1 (PCIF1) to create m6Am. Curiously, the fly ortholog of mammalian PCIF1 is demonstrated to be catalytic-dead, and its functions are not known. Here, we show that Pcif1 mutant flies display a reduced fertility which is particularly marked in females. Deep sequencing analysis of Pcif1 mutant ovaries revealed transcriptome changes with a notable increase in expression of genes belonging to the mitochondrial ATP synthetase complex. Furthermore, the Pcif1 protein is distributed along euchromatic regions of polytene chromosomes, and the Pcif1 mutation behaved as a modifier of position-effect-variegation (PEV) suppressing the heterochromatin-dependent silencing of the white gene. Similar or stronger changes in the transcriptome and PEV phenotype were observed in flies that expressed a cytosolic version of Pcif1. These results point to a nuclear cotranscriptional gene regulatory role for the catalytic-dead fly Pcif1 that is probably based on its conserved ability to interact with the RNA polymerase II carboxy-terminal domain.
Keywords: N6-methyladenosine; Pcif1; m6A; m6Am; position-effect variegation (PEV); transcription.
© 2023 Franco et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Boulias K, Toczydlowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, Guez T, Vasseur JJ, Debart F, Aravind L, et al. 2019. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol Cell 75: 631–643.e638. 10.1016/j.molcel.2019.06.006 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
