Functional Changes in GABA and Glutamate during Motor Learning
- PMID: 36754626
- PMCID: PMC9961379
- DOI: 10.1523/ENEURO.0356-20.2023
Functional Changes in GABA and Glutamate during Motor Learning
Abstract
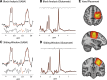
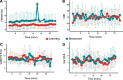
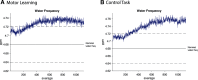
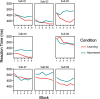
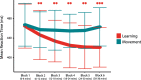
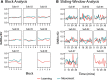
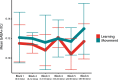
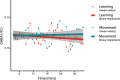
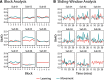
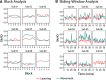
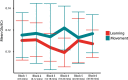
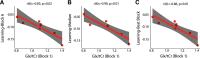
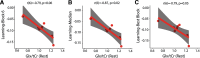
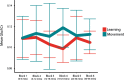
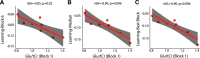
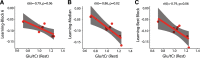
Functional magnetic resonance spectroscopy (fMRS) of GABA at 3 T poses additional challenges compared with fMRS of other metabolites because of the difficulties of measuring GABA levels; GABA is present in the brain at relatively low concentrations, and its signal is overlapped by higher concentration metabolites. Using 7 T fMRS, GABA levels have been shown to decrease specifically during motor learning (and not during a control task). Though the use of 7 T is appealing, access is limited. For GABA fMRS to be widely accessible, it is essential to develop this method at 3 T. Nine healthy right-handed participants completed a motor learning and a control button-pressing task. fMRS data were acquired from the left sensorimotor cortex during the task using a continuous GABA-edited MEGA-PRESS acquisition at 3 T. We found no significant changes in GABA+/tCr, Glx/tCr, or Glu/tCr levels in either task; however, we show a positive relationship between motor learning and glutamate levels both at rest and at the start of the task. Though further refinement and validation of this method is needed, this study represents a further step in using fMRS at 3 T to probe GABA levels in both healthy cognition and clinical disorders.
Keywords: GABA; GABA editing; MEGA-PRESS; functional magnetic resonance spectroscopy (fMRS); glutamate; motor learning.
Copyright © 2023 Bell et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

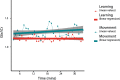
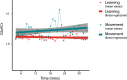
References
-
- Bell T, Goerzen D, Near J, Harris AD (2022) A comparison of human brain GABA levels measured at 3T and 7T. In: Joint annual meeting ISMRM-ESMRMB & ISMRT 31st annual meeting, p 1893. London: ISMRM-ESMRMB & ISMRT.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
