VGLL3 is a mechanosensitive protein that promotes cardiac fibrosis through liquid-liquid phase separation
- PMID: 36754961
- PMCID: PMC9908974
- DOI: 10.1038/s41467-023-36189-6
VGLL3 is a mechanosensitive protein that promotes cardiac fibrosis through liquid-liquid phase separation
Abstract
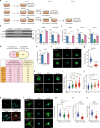
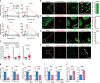
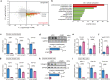
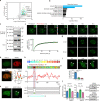
Myofibroblasts cause tissue fibrosis by producing extracellular matrix proteins, such as collagens. Humoral factors like TGF-β, and matrix stiffness are important for collagen production by myofibroblasts. However, the molecular mechanisms regulating their ability to produce collagen remain poorly characterised. Here, we show that vestigial-like family member 3 (VGLL3) is specifically expressed in myofibroblasts from mouse and human fibrotic hearts and promotes collagen production. Further, substrate stiffness triggers VGLL3 translocation into the nucleus through the integrin β1-Rho-actin pathway. In the nucleus, VGLL3 undergoes liquid-liquid phase separation via its low-complexity domain and is incorporated into non-paraspeckle NONO condensates containing EWS RNA-binding protein 1 (EWSR1). VGLL3 binds EWSR1 and suppresses miR-29b, which targets collagen mRNA. Consistently, cardiac fibrosis after myocardial infarction is significantly attenuated in Vgll3-deficient mice, with increased miR-29b expression. Overall, our results reveal an unrecognised VGLL3-mediated pathway that controls myofibroblasts' collagen production, representing a novel therapeutic target for tissue fibrosis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

