Human Satellite 1A analysis provides evidence of pericentromeric transcription
- PMID: 36755311
- PMCID: PMC9909926
- DOI: 10.1186/s12915-023-01521-5
Human Satellite 1A analysis provides evidence of pericentromeric transcription
Abstract
Background: Pericentromeric regions of human chromosomes are composed of tandem-repeated and highly organized sequences named satellite DNAs. Human classical satellite DNAs are classified into three families named HSat1, HSat2, and HSat3, which have historically posed a challenge for the assembly of the human reference genome where they are misrepresented due to their repetitive nature. Although being known for a long time as the most AT-rich fraction of the human genome, classical satellite HSat1A has been disregarded in genomic and transcriptional studies, falling behind other human satellites in terms of functional knowledge. Here, we aim to characterize and provide an understanding on the biological relevance of HSat1A.
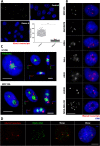
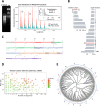
Results: The path followed herein trails with HSat1A isolation and cloning, followed by in silico analysis. Monomer copy number and expression data was obtained in a wide variety of human cell lines, with greatly varying profiles in tumoral/non-tumoral samples. HSat1A was mapped in human chromosomes and applied in in situ transcriptional assays. Additionally, it was possible to observe the nuclear organization of HSat1A transcripts and further characterize them by 3' RACE-Seq. Size-varying polyadenylated HSat1A transcripts were detected, which possibly accounts for the intricate regulation of alternative polyadenylation.
Conclusion: As far as we know, this work pioneers HSat1A transcription studies. With the emergence of new human genome assemblies, acrocentric pericentromeres are becoming relevant characters in disease and other biological contexts. HSat1A sequences and associated noncoding RNAs will most certainly prove significant in the future of HSat research.
Keywords: HSat1A; Noncoding RNA; Pericentromere; Satellite transcription; Transcript polyadenylation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests .
Figures


References
-
- Yunis JJ, Yasmineh WG. Heterochromatin, satellite DNA, and cell function. Science. 1971;174(4015):1200–1209. - PubMed
-
- Kit S. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol. 1961;3(6):711–IN712. - PubMed
-
- Choo KA. The centromere, vol. 320. Oxford: Oxford University Press; 1997.
-
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum Genet. 1997;100(3–4):291–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

