Mutational characterization of Omicron SARS-CoV-2 lineages circulating in Chhattisgarh, a central state of India
- PMID: 36755883
- PMCID: PMC9899822
- DOI: 10.3389/fmed.2022.1082846
Mutational characterization of Omicron SARS-CoV-2 lineages circulating in Chhattisgarh, a central state of India
Abstract
Introduction: The emergence of the Omicron SARS-CoV-2 variant from various states of India in early 2022 has caused fear of its rapid spread. The lack of such reports from Chhattisgarh (CG), a central state in India, has prompted us to identify the Omicron circulating lineages and their mutational dynamics.
Materials and methods: Whole-genome sequencing (WGS) of SARS-CoV-2 was performed in 108 SARS-CoV-2 positive combined samples of nasopharyngeal and oropharyngeal swabs obtained from an equal number of patients.
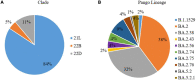
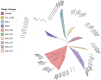
Results: All 108 SARS-CoV-2 sequences belonged to Omicron of clade 21L (84%), 22B (11%), and 22D (5%). BA.2 and its sub-lineages were predominantly found in 93.5% of patients, BA.5.2 and its sub-lineage BA.5.2.1 in 4.6% of patients, and B.1.1.529 in 2% of patients. Various BA.2 sub-lineages identified were BA.2 (38%), BA.2.38 (32%), BA.2.75 (9.25%), BA.2.56, BA.2.76, and BA.5.2.1 (5% each), BA.2.74 (4.6%), BA.5.2.1 (3.7%), BA.2.43 and B.1.1.529 (1.8% each), and BA.5.2 (0.9%). Maximum mutations were noticed in the spike (46), followed by the nucleocapsid (5), membrane (3), and envelope (2) genes. Mutations detected in the spike gene of different Omicron variants were BA.1.1.529 (32), BA.2 (44), BA.2.38 (37), BA.2.43 (38), BA.2.56 (30), BA.2.74 (31), BA.2.75 (37), BA.2.76 (32), BA.5.2, and BA.5.2.1 (38 similar mutations). The spike gene showed the signature mutations of T19I and V213G in the N-terminal domain (NTD), S373P, S375F, T376A, and D405N in receptor-binding domain (RBD), D614G, H655Y, N679K, and P681H at the furin cleavage site, N764K and D796K in fusion peptide, and Q954H and N969K in heptapeptide repeat sequence (HR)1. Notably, BA.2.43 exhibited a novel mutation of E1202Q in the C terminal. Other sites included ORF1a harboring 13 mutations followed by ORF1b (6), ORF3a (2), and ORF6 and ORF8 (1 mutation each).
Conclusion: BA.2 followed by BA.2.38 was the predominant Omicron lineage circulating in Chhattisgarh. BA.2.75 could supersede other Omicron due to its mutational consortium advantage. The periodical genomic monitoring of Omicron variants is thus required for real-time assessment of circulating strains and their mutational-induced severity.
Keywords: BA.2.38; BA.2.75; BA.5; COVID-19; Omicron; SARS-CoV-2.
Copyright © 2023 Singh, Sharma, Shaw, Bhargava and Negi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Characterization of the viral genome of Omicron variants of SARS-CoV-2 circulating in Tripura, a remote frontier state in Northeastern India.Mol Biol Rep. 2024 Oct 28;51(1):1100. doi: 10.1007/s11033-024-10048-z. Mol Biol Rep. 2024. PMID: 39466467
-
Mutational Analysis of Circulating Omicron SARS-CoV-2 Lineages in the Al-Baha Region of Saudi Arabia.J Multidiscip Healthc. 2023 Jul 27;16:2117-2136. doi: 10.2147/JMDH.S419859. eCollection 2023. J Multidiscip Healthc. 2023. PMID: 37529147 Free PMC article.
-
Alpha to JN.1 variants: SARS-CoV-2 genomic analysis unfolding its various lineages/sublineages evolved in Chhattisgarh, India from 2020 to 2024.World J Virol. 2025 Jun 25;14(2):100001. doi: 10.5501/wjv.v14.i2.100001. World J Virol. 2025. PMID: 40575649 Free PMC article.
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
-
The emergence of SARS-CoV-2 Omicron subvariants: current situation and future trends.Infez Med. 2022 Dec 1;30(4):480-494. doi: 10.53854/liim-3004-2. eCollection 2022. Infez Med. 2022. PMID: 36482957 Free PMC article. Review.
Cited by
-
Genomic characterization of Influenza A (H1N1)pdm09 and SARS-CoV-2 from Influenza Like Illness (ILI) and Severe Acute Respiratory Illness (SARI) cases reported between July-December, 2022.Sci Rep. 2024 May 9;14(1):10660. doi: 10.1038/s41598-024-58993-w. Sci Rep. 2024. PMID: 38724525 Free PMC article.
-
Characterization of the viral genome of Omicron variants of SARS-CoV-2 circulating in Tripura, a remote frontier state in Northeastern India.Mol Biol Rep. 2024 Oct 28;51(1):1100. doi: 10.1007/s11033-024-10048-z. Mol Biol Rep. 2024. PMID: 39466467
-
CAR T-cell immunotherapy as the next horizon in cancer eradication: current landscape, challenges, and future directions.Med Oncol. 2025 Aug 6;42(9):410. doi: 10.1007/s12032-025-02957-1. Med Oncol. 2025. PMID: 40770153 Review.
-
AnnCovDB: a manually curated annotation database for mutations in SARS-CoV-2 spike protein.Database (Oxford). 2025 Feb 12;2025:baaf002. doi: 10.1093/database/baaf002. Database (Oxford). 2025. PMID: 39937661 Free PMC article.
-
Clinical and humoral immune response characterization of SARS-CoV-2 Omicron BA.2.38 infection in pediatric patients.Heliyon. 2023 Jul 11;9(7):e18093. doi: 10.1016/j.heliyon.2023.e18093. eCollection 2023 Jul. Heliyon. 2023. PMID: 37519697 Free PMC article.
References
-
- WHO. Tracking SARS-CoV-2 Variants. (2022). Available online at: https://www.who.int/activities/tracking-SARSCoV-2-variants (accessed June 21, 2022).
-
- Khandia R, Singhal S, Alqahtani T, Kamal M, El-Shall N, Nainu F, et al. Emergence of SARS-CoV-2 omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ Res. (2022) 209:112816. 10.1016/j.envres.2022.112816 - DOI - PMC - PubMed
-
- Singh P, Sharma K, Singh P, Bhargava A, Negi S, Sharma P, et al. Genomic characterization unravelling the causative role of SARS-CoV-2 delta variant of lineage B.1.617.2 in 2nd wave of COVID-19 pandemic in Chhattisgarh. India Microb Pathog. (2022) 164:105404. 10.1016/j.micpath.2022.105404 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

