Community diversity is associated with intra-species genetic diversity and gene loss in the human gut microbiome
- PMID: 36757364
- PMCID: PMC9977275
- DOI: 10.7554/eLife.78530
Community diversity is associated with intra-species genetic diversity and gene loss in the human gut microbiome
Abstract
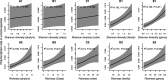
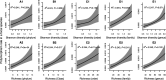
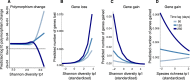
How the ecological process of community assembly interacts with intra-species diversity and evolutionary change is a longstanding question. Two contrasting hypotheses have been proposed: Diversity Begets Diversity (DBD), in which taxa tend to become more diverse in already diverse communities, and Ecological Controls (EC), in which higher community diversity impedes diversification. Previously, using 16S rRNA gene amplicon data across a range of microbiomes, we showed a generally positive relationship between taxa diversity and community diversity at higher taxonomic levels, consistent with the predictions of DBD (Madi et al., 2020). However, this positive 'diversity slope' plateaus at high levels of community diversity. Here we show that this general pattern holds at much finer genetic resolution, by analyzing intra-species strain and nucleotide variation in static and temporally sampled metagenomes from the human gut microbiome. Consistent with DBD, both intra-species polymorphism and strain number were positively correlated with community Shannon diversity. Shannon diversity is also predictive of increases in polymorphism over time scales up to ~4-6 months, after which the diversity slope flattens and becomes negative - consistent with DBD eventually giving way to EC. Finally, we show that higher community diversity predicts gene loss at a future time point. This observation is broadly consistent with the Black Queen Hypothesis, which posits that genes with functions provided by the community are less likely to be retained in a focal species' genome. Together, our results show that a mixture of DBD, EC, and Black Queen may operate simultaneously in the human gut microbiome, adding to a growing body of evidence that these eco-evolutionary processes are key drivers of biodiversity and ecosystem function.
Keywords: Black Queen; ecology; evolution; evolutionary biology; metagenomics; microbiome; none; population genetics.
© 2023, Madi et al.
Conflict of interest statement
NM, DC, RW, BS, NG No competing interests declared
Figures






Update of
References
-
- Bolker B. Mixedmodels-misc. swh:1:rev:3b8b732667b6a261a3384f5cade8e78b68230f3aSoftware Heritage. 2023 https://archive.softwareheritage.org/swh:1:dir:8e83cdf33e3ea4f240774111d...
-
- Brooks ME, Kristensen K, van Benthem KJ, Magnusson A, Berg CW, Nielsen A, Skaug HJ, Mächler M, Bolker BM. Modeling Zero-Inflated Count Data with GlmmTMB. bioRxiv. 2017 doi: 10.1101/132753. - DOI

