Conserved chloroplast genome sequences of the genus Clerodendrum Linn. (Lamiaceae) as a super-barcode
- PMID: 36757949
- PMCID: PMC9910634
- DOI: 10.1371/journal.pone.0277809
Conserved chloroplast genome sequences of the genus Clerodendrum Linn. (Lamiaceae) as a super-barcode
Abstract
Background: The plants of the genus Clerodendrum L. have great potential for development as an ornamental and important herbal resource. There is no significant morphological difference among many species of the genus Clerodendrum, which will lead to confusion among the herbs of this genus and ultimately affect the quality of the herbs. The chloroplast genome will contribute to the development of new markers used for the identification and classification of species.
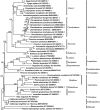
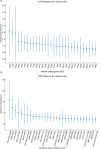
Methods and results: Here, we obtained the complete chloroplast genome sequences of Clerodendrum chinense (Osbeck) Mabberley and Clerodendrum thomsoniae Balf.f. using the next generation DNA sequencing technology. The chloroplast genomes of the two species all encode a total of 112 unique genes, including 80 protein-coding, 28 tRNA, and four rRNA genes. A total of 44-42 simple sequence repeats, 19-16 tandem repeats and 44-44 scattered repetitive sequences were identified. Phylogenetic analyses showed that the nine Clerodendrum species were classified into two clades and together formed a monophyletic group. Selective pressure analyses of 77 protein-coding genes showed that there was no gene under positive selection in the Clerodendrum branch. Analyses of sequence divergence found two intergenic regions: trnH-GUG-psbA, nhdD-psaC, exhibiting a high degree of variations. Meanwhile, there was no hypervariable region identified in protein coding genes. However, the sequence identities of these two intergenic spacers (IGSs) are greater than 99% among some species, which will result in the two IGSs not being used to distinguish Clerodendrum species. Analysis of the structure at the LSC (Large single copy) /IR (Inverted repeat) and SSC (Small single copy)/IR boundary regions showed dynamic changes. The above results showed that the complete chloroplast genomes can be used as a super-barcode to identify these Clerodendrum species. The study lay the foundation for the understanding of the evolutionary process of the genus Clerodendrum.
Copyright: © 2023 Chen et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Chloroplast Genome Structure and Phylogenetic Analysis of 13 Lamiaceae Plants in Tibet.Front Biosci (Landmark Ed). 2023 Jun 8;28(6):110. doi: 10.31083/j.fbl2806110. Front Biosci (Landmark Ed). 2023. PMID: 37395020
-
Complete chloroplast genome sequence and phylogenetic analysis of Clerodendrum japonicum (Thunb.) Sweet (Ajugoideae, Lamiaceae).Mitochondrial DNA B Resour. 2021 Jul 5;6(8):2218-2220. doi: 10.1080/23802359.2021.1944367. eCollection 2021. Mitochondrial DNA B Resour. 2021. PMID: 34263052 Free PMC article.
-
Complete chloroplast genome of Stephania tetrandra (Menispermaceae) from Zhejiang Province: insights into molecular structures, comparative genome analysis, mutational hotspots and phylogenetic relationships.BMC Genomics. 2021 Dec 6;22(1):880. doi: 10.1186/s12864-021-08193-x. BMC Genomics. 2021. PMID: 34872502 Free PMC article.
-
Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes. (Melanthiaceae).Gene. 2013 Nov 10;530(2):229-35. doi: 10.1016/j.gene.2013.07.100. Epub 2013 Aug 23. Gene. 2013. PMID: 23973725
-
Comparative Analysis of the Chloroplast Genomes of Eight Species of the Genus Lirianthe Spach with Its Generic Delimitation Implications.Int J Mol Sci. 2024 Mar 20;25(6):3506. doi: 10.3390/ijms25063506. Int J Mol Sci. 2024. PMID: 38542477 Free PMC article.
Cited by
-
The complete chloroplast genome sequence and phylogenetic analysis of Volkameria inermis Linnaeus 1753 (Lamiaceae), a tropical and subtropical coastal shrub.Mitochondrial DNA B Resour. 2025 Aug 5;10(9):779-783. doi: 10.1080/23802359.2025.2541623. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 40778371 Free PMC article.
-
Plastome comparison and phylogenomics of Chinese endemic Schnabelia (Lamiaceae): insights into plastome evolution and species divergence.BMC Plant Biol. 2025 May 7;25(1):600. doi: 10.1186/s12870-025-06647-y. BMC Plant Biol. 2025. PMID: 40335944 Free PMC article.
-
Comparative analysis of chloroplast genome and new insights into phylogenetic relationships of Ajuga and common adulterants.Front Plant Sci. 2023 Oct 25;14:1251829. doi: 10.3389/fpls.2023.1251829. eCollection 2023. Front Plant Sci. 2023. PMID: 37954994 Free PMC article.
-
Comparative study on chloroplast genomes of three Hansenia forbesii varieties (Apiaceae).PLoS One. 2023 Jun 1;18(6):e0286587. doi: 10.1371/journal.pone.0286587. eCollection 2023. PLoS One. 2023. PMID: 37262084 Free PMC article.
References
-
- Flora of China: Clerodendrum: IBCAS Publishing iPlant; 2021. Available from: http://www.iplant.cn/info/Clerodendrum?t=z.
-
- Shi-qiao L, Fu-jun Z, Xian G, Zhong-yao H, Wen-bin H. Research advance on medicinal plants of Clerodendrum. Drug Evaluation Research. 2011;34(6):469–73.
-
- Nguyen TH, Le HD, Kim TNT, The HP, Nguyen TM, Cornet V, et al.. Anti-Inflammatory and Antioxidant Properties of the Ethanol Extract of Clerodendrum Cyrtophyllum Turcz in Copper Sulfate-Induced Inflammation in Zebrafish. Antioxidants (Basel, Switzerland). 2020;9(3). doi: 10.3390/antiox9030192 - DOI - PMC - PubMed
-
- Fern K. Tropical Plants Database 2022-03-28. Available from: tropical.theferns.info/viewtropical.php?id=Clerodendrum+chinense.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

