Epigenetics as a mediator of plasticity in cancer
- PMID: 36758093
- PMCID: PMC10249049
- DOI: 10.1126/science.aaw3835
Epigenetics as a mediator of plasticity in cancer
Abstract
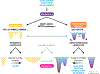
The concept of an epigenetic landscape describing potential cellular fates arising from pluripotent cells, first advanced by Conrad Waddington, has evolved in light of experiments showing nondeterministic outcomes of regulatory processes and mathematical methods for quantifying stochasticity. In this Review, we discuss modern approaches to epigenetic and gene regulation landscapes and the associated ideas of entropy and attractor states, illustrating how their definitions are both more precise and relevant to understanding cancer etiology and the plasticity of cancerous states. We address the interplay between different types of regulatory landscapes and how their changes underlie cancer progression. We also consider the roles of cellular aging and intrinsic and extrinsic stimuli in modulating cellular states and how landscape alterations can be quantitatively mapped onto phenotypic outcomes and thereby used in therapy development.
Figures

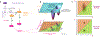
References
-
- Fidler IJ, Tumor Heterogeneity and the Biology of Cancer Invasion and Metastasis1. Cancer Research 38, 2651–2660 (1978). - PubMed
-
- Nowell PC, The Clonal Evolution of Tumor Cell Populations: Acquired genetic lability permits stepwise selection of variant sublines and underlies tumor progression. Science 194, 23–28 (1976). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

