Identification and genome sequencing of an influenza H3N2 variant in wastewater from elementary schools during a surge of influenza A cases in Las Vegas, Nevada
- PMID: 36758698
- PMCID: PMC9909754
- DOI: 10.1016/j.scitotenv.2023.162058
Identification and genome sequencing of an influenza H3N2 variant in wastewater from elementary schools during a surge of influenza A cases in Las Vegas, Nevada
Abstract
Real-time surveillance of infectious diseases at schools or in communities is often hampered by delays in reporting due to resource limitations and infrastructure issues. By incorporating quantitative PCR and genome sequencing, wastewater surveillance has been an effective complement to public health surveillance at the community and building-scale for pathogens such as poliovirus, SARS-CoV-2, and even the monkeypox virus. In this study, we asked whether wastewater surveillance programs at elementary schools could be leveraged to detect RNA from influenza viruses shed in wastewater. We monitored for influenza A and B viral RNA in wastewater from six elementary schools from January to May 2022. Quantitative PCR led to the identification of influenza A viral RNA at three schools, which coincided with the lifting of COVID-19 restrictions and a surge in influenza A infections in Las Vegas, Nevada, USA. We performed genome sequencing of wastewater RNA, leading to the identification of a 2021-2022 vaccine-resistant influenza A (H3N2) 3C.2a1b.2a.2 subclade. We next tested wastewater samples from a treatment plant that serviced the elementary schools, but we were unable to detect the presence of influenza A/B RNA. Together, our results demonstrate the utility of near-source wastewater surveillance for the detection of local influenza transmission in schools, which has the potential to be investigated further with paired school-level influenza incidence data.
Keywords: COVID-19; Elementary schools, mutation; H3N2; Influenza; SARS-CoV-2; Wastewater.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest Dr. Carolina Koutras is an employee of R-Zero Systems.
Figures



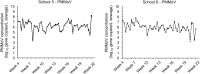
References
-
- Abdool-Ghany A.A., Sahwell P.J., Klaus J., Gidley M.L., Sinigalliano C.D., Solo-Gabriele H.M. Fecal indicator bacteria levels at a marine beach before, during, and after the COVID-19 shutdown period and associations with decomposing seaweed and human presence. Sci. Total Environ. 2022;851 doi: 10.1016/j.scitotenv.2022.158349. - DOI - PubMed
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
-
- Bolton M.J., Ort J.T., McBride R., Swanson N.J., Wilson J., Awofolaju M., Furey C., Greenplate A.R., Drapeau E.M., Pekosz A., Paulson J.C., Hensley S.E. Antigenic and virological properties of an H3N2 variant that continues to dominate the 2021–22 Northern Hemisphere influenza season. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110897. - DOI - PMC - PubMed
-
- Brumfield K.D., Leddy M., Usmani M., Cotruvo J.A., Tien C.-T., Dorsey S., Graubics K., Fanelli B., Zhou I., Registe N., Dadlani M., Wimalarante M., Jinasena D., Abayagunawardena R., Withanachchi C., Huq A., Jutla A., Colwell R.R. Microbiome Analysis for Wastewater Surveillance during COVID-19. mBio. 2022 doi: 10.1128/mbio.00591-22. - DOI - PMC - PubMed
-
- Castro-Gutierrez V., Hassard F., Vu M., Leitao R., Burczynska B., Wildeboer D., Stanton I., Rahimzadeh S., Baio G., Garelick H., Hofman J., Kasprzyk-Hordern B., Kwiatkowska R., Majeed A., Priest S., Grimsley J., Lundy L., Singer A.C., Di Cesare M. Monitoring occurrence of SARS-CoV-2 in school populations: a wastewater-based approach. PLoS One. 2022;17 doi: 10.1371/journal.pone.0270168. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

