The main protease of SARS-CoV-2 cleaves histone deacetylases and DCP1A, attenuating the immune defense of the interferon-stimulated genes
- PMID: 36758802
- PMCID: PMC9907797
- DOI: 10.1016/j.jbc.2023.102990
The main protease of SARS-CoV-2 cleaves histone deacetylases and DCP1A, attenuating the immune defense of the interferon-stimulated genes
Abstract
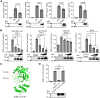
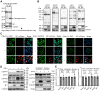
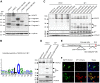
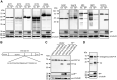
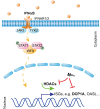
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019, constitutes an emerging human pathogen of zoonotic origin. A critical role in protecting the host against invading pathogens is carried out by interferon-stimulated genes (ISGs), the primary effectors of the type I interferon (IFN) response. All coronaviruses studied thus far have to first overcome the inhibitory effects of the IFN/ISG system before establishing efficient viral replication. However, whether SARS-CoV-2 evades IFN antiviral immunity by manipulating ISG activation remains to be elucidated. Here, we show that the SARS-CoV-2 main protease (Mpro) significantly suppresses the expression and transcription of downstream ISGs driven by IFN-stimulated response elements in a dose-dependent manner, and similar negative regulations were observed in two mammalian epithelial cell lines (simian Vero E6 and human A549). Our analysis shows that to inhibit the ISG production, Mpro cleaves histone deacetylases (HDACs) rather than directly targeting IFN signal transducers. Interestingly, Mpro also abolishes the activity of ISG effector mRNA-decapping enzyme 1a (DCP1A) by cleaving it at residue Q343. In addition, Mpro from different genera of coronaviruses has the protease activity to cleave both HDAC2 and DCP1A, even though the alphacoronaviruse Mpro exhibits weaker catalytic activity in cleaving HDAC2. In conclusion, our findings clearly demonstrate that SARS-CoV-2 Mpro constitutes a critical anti-immune effector that modulates the IFN/ISG system at multiple levels, thus providing a novel molecular explanation for viral immune evasion and allowing for new therapeutic approaches against coronavirus disease 2019 infection.
Keywords: SARS-CoV-2; cleavage; histone deacetylases; interferon-stimulated gene; mRNA-decapping enzyme 1a; main protease.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare there are no conflicts of interest with the contents of this article.
Figures



Similar articles
-
Porcine Deltacoronavirus nsp5 Cleaves DCP1A To Decrease Its Antiviral Activity.J Virol. 2020 Jul 16;94(15):e02162-19. doi: 10.1128/JVI.02162-19. Print 2020 Jul 16. J Virol. 2020. PMID: 32461317 Free PMC article.
-
Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article.
-
Inhibition of the IFN-α JAK/STAT Pathway by MERS-CoV and SARS-CoV-1 Proteins in Human Epithelial Cells.Viruses. 2022 Mar 23;14(4):667. doi: 10.3390/v14040667. Viruses. 2022. PMID: 35458397 Free PMC article.
-
Characterization of SARS-CoV-2 Evasion: Interferon Pathway and Therapeutic Options.Viruses. 2022 Jun 8;14(6):1247. doi: 10.3390/v14061247. Viruses. 2022. PMID: 35746718 Free PMC article. Review.
-
Dysregulated Interferon Response and Immune Hyperactivation in Severe COVID-19: Targeting STATs as a Novel Therapeutic Strategy.Front Immunol. 2022 May 17;13:888897. doi: 10.3389/fimmu.2022.888897. eCollection 2022. Front Immunol. 2022. PMID: 35663932 Free PMC article. Review.
Cited by
-
Allostery in homodimeric SARS-CoV-2 main protease.Commun Biol. 2024 Nov 4;7(1):1435. doi: 10.1038/s42003-024-07138-w. Commun Biol. 2024. PMID: 39496839 Free PMC article.
-
Swine acute diarrhoea syndrome coronavirus (SADS-CoV) Nsp5 antagonizes type I interferon signaling by cleaving DCP1A.Front Immunol. 2023 May 22;14:1196031. doi: 10.3389/fimmu.2023.1196031. eCollection 2023. Front Immunol. 2023. PMID: 37283741 Free PMC article.
-
Research Progress on the Structure and Function, Immune Escape Mechanism, Antiviral Drug Development Methods, and Clinical Use of SARS-CoV-2 Mpro.Molecules. 2025 Jan 16;30(2):351. doi: 10.3390/molecules30020351. Molecules. 2025. PMID: 39860219 Free PMC article. Review.
-
The nonstructural protein 13 of porcine deltacoronavirus coordinates ATP-driven duplex unwinding and ATP-independent strand annealing for nucleic acid remodeling.BMC Vet Res. 2025 Jul 2;21(1):414. doi: 10.1186/s12917-025-04851-4. BMC Vet Res. 2025. PMID: 40597348 Free PMC article.
-
Amino acid T25 in the substrate-binding domain of SARS-CoV-2 nsp5 is involved in viral replication in the mouse lung.PLoS One. 2024 Dec 6;19(12):e0312800. doi: 10.1371/journal.pone.0312800. eCollection 2024. PLoS One. 2024. PMID: 39642113 Free PMC article.
References
-
- Pormohammad A., Ghorbani S., Baradaran B., Khatami A., R J.T., Mansournia M.A., et al. Clinical characteristics, laboratory findings, radiographic signs and outcomes of 61,742 patients with confirmed COVID-19 infection: a systematic review and meta-analysis. Microb. Pathog. 2020;147:104390. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

