Prioritization of Drug Targets for Neurodegenerative Diseases by Integrating Genetic and Proteomic Data From Brain and Blood
- PMID: 36759259
- PMCID: PMC12272336
- DOI: 10.1016/j.biopsych.2022.11.002
Prioritization of Drug Targets for Neurodegenerative Diseases by Integrating Genetic and Proteomic Data From Brain and Blood
Abstract
Background: Neurodegenerative diseases are among the most prevalent and devastating neurological disorders, with few effective prevention and treatment strategies. We aimed to integrate genetic and proteomic data to prioritize drug targets for neurodegenerative diseases.
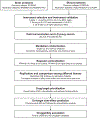
Methods: We screened human proteomes through Mendelian randomization to identify causal mediators of Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis, multiple sclerosis, frontotemporal dementia, and Lewy body dementia. For instruments, we used brain and blood protein quantitative trait loci identified from one genome-wide association study with 376 participants and another with 3301 participants, respectively. Causal associations were subsequently validated by sensitivity analyses and colocalization. The safety and druggability of identified targets were also evaluated.
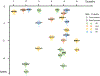
Results: Our analyses showed targeting BIN1, GRN, and RET levels in blood as well as ACE, ICA1L, MAP1S, SLC20A2, and TOM1L2 levels in brain might reduce Alzheimer's disease risk, while ICA1L, SLC20A2, and TOM1L2 were not recommended as prioritized drugs due to the identified potential side effects. Brain CD38, DGKQ, GPNMB, and SEC23IP were candidate targets for Parkinson's disease. Among them, GPNMB was the most promising target for Parkinson's disease with their causal relationship evidenced by studies on both brain and blood tissues. Interventions targeting FCRL3, LMAN2, and MAPK3 in blood and DHRS11, FAM120B, SHMT1, and TSFM in brain might affect multiple sclerosis risk. The risk of amyotrophic lateral sclerosis might be reduced by medications targeting DHRS11, PSMB3, SARM1, and SCFD1 in brain.
Conclusions: Our study prioritized 22 proteins as targets for neurodegenerative diseases and provided preliminary evidence for drug development. Further studies are warranted to validate these targets.
Keywords: Drug discovery; Genetics; Mendelian randomization; Neurodegenerative disease; Omics; Proteomics.
Copyright © 2022 Society of Biological Psychiatry. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DISCLOSURES
GWAS summary statistics of QTLs and neurodegenerative diseases are available from the peer-reviewed articles or the corresponding authors on request. Other phenotypes of potential side effects are available from the MRC IEU OpenGWAS Project (
The authors report no biomedical financial interests or potential conflicts of interest.
Figures


Comment in
-
Proteomic Data Advance Targeted Drug Development for Neurogenerative Diseases.Biol Psychiatry. 2023 May 1;93(9):754-755. doi: 10.1016/j.biopsych.2023.02.003. Biol Psychiatry. 2023. PMID: 37045511 Free PMC article. No abstract available.
References
-
- Villoslada P, Baeza-Yates R, Masdeu JC (2020): Reclassifying neurodegenerative diseases. Nat Biomed Eng 4:759–760. - PubMed
-
- The Lancet Neurology (2013): Joining forces to fight neurodegenerative diseases. Lancet Neurol 12:119. - PubMed
-
- Alteri E, Guizzaro L (2018): Be open about drug failures to speed up research. Nature 563:317–319. - PubMed
-
- Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. (2015): The support of human genetic evidence for approved drug indications. Nat Genet 47:856–860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AG079850/AG/NIA NIH HHS/United States
- MR/T033371/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00030/14/MRC_/Medical Research Council/United Kingdom
- MC_U105597119/MRC_/Medical Research Council/United Kingdom
- MR/M008983/1/MRC_/Medical Research Council/United Kingdom
- G0301152/MRC_/Medical Research Council/United Kingdom
- R01 MH120794/MH/NIMH NIH HHS/United States
- MR/M023664/1/MRC_/Medical Research Council/United Kingdom
- R01 AG062268/AG/NIA NIH HHS/United States
- MR/T046015/1/MRC_/Medical Research Council/United Kingdom
- MR/M008525/1/MRC_/Medical Research Council/United Kingdom
- MR/L023784/2/MRC_/Medical Research Council/United Kingdom
- MC_UU_00005/12/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

