Cross-protection and cross-feeding between Klebsiella pneumoniae and Acinetobacter baumannii promotes their co-existence
- PMID: 36759602
- PMCID: PMC9911699
- DOI: 10.1038/s41467-023-36252-2
Cross-protection and cross-feeding between Klebsiella pneumoniae and Acinetobacter baumannii promotes their co-existence
Abstract
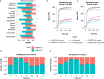
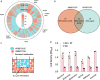
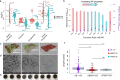
Acinetobacter baumannii and Klebsiella pneumoniae are opportunistic pathogens frequently co-isolated from polymicrobial infections. The infections where these pathogens co-exist can be more severe and recalcitrant to therapy than infections caused by either species alone, however there is a lack of knowledge on their potential synergistic interactions. In this study we characterise the genomes of A. baumannii and K. pneumoniae strains co-isolated from a single human lung infection. We examine various aspects of their interactions through transcriptomic, phenomic and phenotypic assays that form a basis for understanding their effects on antimicrobial resistance and virulence during co-infection. Using co-culturing and analyses of secreted metabolites, we discover the ability of K. pneumoniae to cross-feed A. baumannii by-products of sugar fermentation. Minimum inhibitory concentration testing of mono- and co-cultures reveals the ability for A. baumannii to cross-protect K. pneumoniae against the cephalosporin, cefotaxime. Our study demonstrates distinct syntrophic interactions occur between A. baumannii and K. pneumoniae, helping to elucidate the basis for their co-existence in polymicrobial infections.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Coexistence of Klebsiella pneumoniae and Acinetobacter baumannii contributes to antibiotic resistance.Int J Antimicrob Agents. 2023 Nov;62(5):106993. doi: 10.1016/j.ijantimicag.2023.106993. Epub 2023 Oct 2. Int J Antimicrob Agents. 2023. PMID: 37793556 No abstract available.
References
-
- Eijkelkamp BA, et al. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol. Lett. 2011;323:44–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

