ApoE4 associated with severe COVID-19 outcomes via downregulation of ACE2 and imbalanced RAS pathway
- PMID: 36759834
- PMCID: PMC9910247
- DOI: 10.1186/s12967-023-03945-7
ApoE4 associated with severe COVID-19 outcomes via downregulation of ACE2 and imbalanced RAS pathway
Abstract
Background: Recent numerous epidemiology and clinical association studies reported that ApoE polymorphism might be associated with the risk and severity of coronavirus disease 2019 (COVID-19), and yielded inconsistent results. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection relies on its spike protein binding to angiotensin-converting enzyme 2 (ACE2) receptor expressed on host cell membranes.
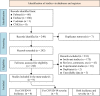
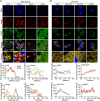
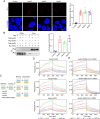
Methods: A meta-analysis was conducted to clarify the association between ApoE polymorphism and the risk and severity of COVID-19. Multiple protein interaction assays were utilized to investigate the potential molecular link between ApoE and the SARS-CoV-2 primary receptor ACE2, ApoE and spike protein. Immunoblotting and immunofluorescence staining methods were used to access the regulatory effect of different ApoE isoform on ACE2 protein expression.
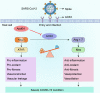
Results: ApoE gene polymorphism (ε4 carrier genotypes VS non-ε4 carrier genotypes) is associated with the increased risk (P = 0.0003, OR = 1.44, 95% CI 1.18-1.76) and progression (P < 0.00001, OR = 1.85, 95% CI 1.50-2.28) of COVID-19. ApoE interacts with both ACE2 and the spike protein but did not show isoform-dependent binding effects. ApoE4 significantly downregulates ACE2 protein expression in vitro and in vivo and subsequently decreases the conversion of Ang II to Ang 1-7.
Conclusions: ApoE4 increases SARS-CoV-2 infectivity in a manner that may not depend on differential interactions with the spike protein or ACE2. Instead, ApoE4 downregulates ACE2 protein expression and subsequently the dysregulation of renin-angiotensin system (RAS) may provide explanation by which ApoE4 exacerbates COVID-19 disease.
Keywords: ACE2; ApoE4; COVID-19; SARS-CoV-2; Spike.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

