Endocrine modulation of brain-skeleton axis driven by neural stem cell-derived perilipin 5 in the lipid metabolism homeostasis for bone regeneration
- PMID: 36760127
- PMCID: PMC10188646
- DOI: 10.1016/j.ymthe.2023.02.004
Endocrine modulation of brain-skeleton axis driven by neural stem cell-derived perilipin 5 in the lipid metabolism homeostasis for bone regeneration
Abstract
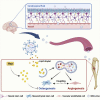
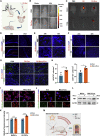
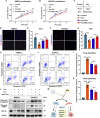
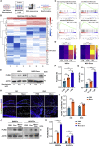
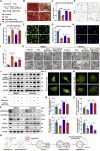
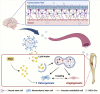
Factors released from the nervous system always play crucial roles in modulating bone metabolism and regeneration. How the brain-driven endocrine axes maintain bone homeostasis, especially under metabolic disorders, remains obscure. Here, we found that neural stem cells (NSCs) residing in the subventricular zone participated in lipid metabolism homeostasis of regenerative bone through exosomal perilipin 5 (PLIN5). Fluorescence-labeled exosomes tracing and histological detection identified that NSC-derived exosomes (NSC-Exo) could travel from the lateral ventricle into bone injury sites. Homocysteine (Hcy) led to osteogenic and angiogenic impairment, whereas the NSC-Exo were confirmed to restore it. Mecobalamin, a clinically used neurotrophic drug, further enhanced the protective effects of NSC-Exo through increased PLIN5 expression. Mechanistically, NSC-derived PLIN5 reversed excessive Hcy-induced lipid metabolic imbalance and aberrant lipid droplet accumulation through lipophagy-dependent intracellular lipolysis. Intracerebroventricular administration of mecobalamin and/or AAV-shPlin5 confirmed the effects of PLIN5-driven endocrine modulations on new bone formation and vascular reconstruction in hyperhomocysteinemic and high-fat diet models. This study uncovered a novel brain-skeleton axis that NSCs in the mammalian brain modulated bone regeneration through PLIN5-driven lipid metabolism modulation, providing evidence for lipid- or bone-targeted medicine development.
Keywords: bone regeneration; homocysteine; lipid metabolism; mecobalamin; neural stem cell; perilipin 5.
Copyright © 2023 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Kong L.C., Li H.A., Kang Q.L., Li G. An update to the advances in understanding distraction histogenesis: from biological mechanisms to novel clinical applications. J. Orthop. Translat. 2020;25:3–10.
-
- Li D., Chen K., Tang H., Hu S., Xin L., Jing X., He Q., Wang S., Song J., Mei L., et al. A logic-based diagnostic and therapeutic hydrogel with multistimuli responsiveness to orchestrate diabetic bone regeneration. Adv. Mater. 2022;34:e2108430. - PubMed
-
- Loeffler J., Duda G.N., Sass F.A., Dienelt A. The metabolic microenvironment steers bone tissue regeneration. Trends Endocrinol. Metab. 2018;29:99–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

