Genome sequencing identifies complex structural MLH1 variant in unsolved Lynch syndrome
- PMID: 36760167
- PMCID: PMC10265068
- DOI: 10.1002/mgg3.2151
Genome sequencing identifies complex structural MLH1 variant in unsolved Lynch syndrome
Abstract
Background: Lynch syndrome is one of the most common cancer predisposition syndromes. It is caused by inherited changes in the mismatch repair pathway. With current diagnostic approaches, a causative genetic variant can be found in less than 50% of cases. A correct diagnosis is important for ensuring that an appropriate surveillance program is used and that additional high-risk family members are identified.
Methods: We used clinical genome sequencing on DNA from blood and subsequent transcriptome sequencing for confirmation. Data were analyzed using the megSAP pipeline and classified according to basic criteria in diagnostic laboratories. Segregation analyses in family members were conducted via breakpoint PCR.
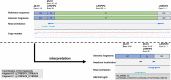
Results: We present a family with the clinical diagnosis of Lynch syndrome in which standard diagnostic tests, such as panel or exome sequencing, were unable to detect the underlying genetic variant. Genome sequencing in the index patient confirmed the previous diagnostic results and identified an additional complex rearrangement with intronic breakpoints involving MLH1 and its neighboring gene LRRFIP2. The previously undetected structural variant was classified as medically relevant. Segregation analysis in the family identified additional at-risk individuals which were offered intensified cancer screening.
Discussion and conclusions: This case illustrates the advantages of clinical genome sequencing in detecting structural variants compared with current diagnostic approaches. Although structural variants are rare in Lynch syndrome families, they seem to be underreported, in part because of technical challenges. Clinical genome sequencing offers a comprehensive genetic characterization detecting a wide range of genetic variants.
Keywords: MLH1; HNPCC; Lynch syndrome; copy-number neutral; genome-sequencing; structural variant.
© 2023 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
There are no competing interests.
Figures

References
-
- Baudhuin, L. M. , Ferber, M. J. , Winters, J. L. , Steenblock, K. J. , Swanson, R. L. , French, A. J. , Butz, M. L. , & Thibodeau, S. N. (2005). Characterization of hMLH1 and hMSH2 gene dosage alterations in Lynch syndrome patients. Gastroenterology, 129(3), 846–854. 10.1053/j.gastro.2005.06.026 - DOI - PubMed
-
- Blaker, H. , Haupt, S. , Morak, M. , Holinski‐Feder, E. , Arnold, A. , Horst, D. , Sieber‐Frank, J. , Seidler, F. , von Winterfeld, M. , Alwers, E. , Chang‐Claude, J. , Brenner, H. , Roth, W. , Engel, C. , Loffler, M. , Moslein, G. , Schackert, H. K. , Weitz, J. , Perne, C. , … German Consortium for Familial Intestinal Cancer . (2020). Age‐dependent performance of BRAF mutation testing in Lynch syndrome diagnostics. International Journal of Cancer, 147(10), 2801–2810. 10.1002/ijc.33273 - DOI - PubMed
-
- Casey, G. , Lindor, N. M. , Papadopoulos, N. , Thibodeau, S. N. , Moskow, J. , Steelman, S. , Buzin, C. H. , Sommer, S. S. , Collins, C. E. , Butz, M. , Aronson, M. , Gallinger, S. , Barker, M. A. , Young, J. P. , Jass, J. R. , Hopper, J. L. , Diep, A. , Bapat, B. , Salem, M. , … Colon Cancer Family Registry . (2005). Conversion analysis for mutation detection in MLH1 and MSH2 in patients with colorectal cancer. JAMA, 293(7), 799–809. 10.1001/jama.293.7.799 - DOI - PMC - PubMed
-
- Cheng, C. , Zhou, Y. , Li, H. , Xiong, T. , Li, S. , Bi, Y. , Kong, P. , Wang, F. , Cui, H. , Li, Y. , Fang, X. , Yan, T. , Li, Y. , Wang, J. , Yang, B. , Zhang, L. , Jia, Z. , Song, B. , Hu, X. , … Cui, Y. (2016). Whole‐genome sequencing reveals diverse models of structural variations in esophageal squamous cell carcinoma. American Journal of Human Genetics, 98(2), 256–274. 10.1016/j.ajhg.2015.12.013 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

