Tetratricopeptide repeat protein SlREC2 positively regulates cold tolerance in tomato
- PMID: 36760172
- PMCID: PMC10152682
- DOI: 10.1093/plphys/kiad085
Tetratricopeptide repeat protein SlREC2 positively regulates cold tolerance in tomato
Abstract
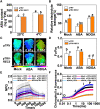
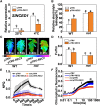
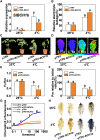
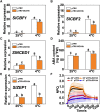
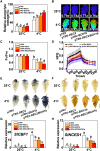
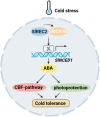
Cold stress is a key environmental constraint that dramatically affects the growth, productivity, and quality of tomato (Solanum lycopersicum); however, the underlying molecular mechanisms of cold tolerance remain poorly understood. In this study, we identified REDUCED CHLOROPLAST COVERAGE 2 (SlREC2) encoding a tetratricopeptide repeat protein that positively regulates tomato cold tolerance. Disruption of SlREC2 largely reduced abscisic acid (ABA) levels, photoprotection, and the expression of C-REPEAT BINDING FACTOR (CBF)-pathway genes in tomato plants under cold stress. ABA deficiency in the notabilis (not) mutant, which carries a mutation in 9-CIS-EPOXYCAROTENOID DIOXYGENASE 1 (SlNCED1), strongly inhibited the cold tolerance of SlREC2-silenced plants and empty vector control plants and resulted in a similar phenotype. In addition, foliar application of ABA rescued the cold tolerance of SlREC2-silenced plants, which confirms that SlNCED1-mediated ABA accumulation is required for SlREC2-regulated cold tolerance. Strikingly, SlREC2 physically interacted with β-RING CAROTENE HYDROXYLASE 1b (SlBCH1b), a key regulatory enzyme in the xanthophyll cycle. Disruption of SlBCH1b severely impaired photoprotection, ABA accumulation, and CBF-pathway gene expression in tomato plants under cold stress. Taken together, this study reveals that SlREC2 interacts with SlBCH1b to enhance cold tolerance in tomato via integration of SlNCED1-mediated ABA accumulation, photoprotection, and the CBF-pathway, thus providing further genetic knowledge for breeding cold-resistant tomato varieties.
© American Society of Plant Biologists 2023. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare no conflict of interest.
Figures



Similar articles
-
Phytochrome A and B Function Antagonistically to Regulate Cold Tolerance via Abscisic Acid-Dependent Jasmonate Signaling.Plant Physiol. 2016 Jan;170(1):459-71. doi: 10.1104/pp.15.01171. Epub 2015 Nov 2. Plant Physiol. 2016. PMID: 26527654 Free PMC article.
-
A transcriptional regulation of ERF15 contributes to ABA-mediated cold tolerance in tomato.Plant Cell Environ. 2024 Apr;47(4):1334-1347. doi: 10.1111/pce.14816. Epub 2024 Jan 14. Plant Cell Environ. 2024. PMID: 38221812
-
The functional analysis of SlNCED1 in tomato pollen development.Cell Mol Life Sci. 2018 Sep;75(18):3457-3472. doi: 10.1007/s00018-018-2809-9. Epub 2018 Apr 9. Cell Mol Life Sci. 2018. PMID: 29632966 Free PMC article.
-
Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato.Plant Physiol. 2012 Jan;158(1):283-98. doi: 10.1104/pp.111.186866. Epub 2011 Nov 22. Plant Physiol. 2012. PMID: 22108525 Free PMC article.
-
Cold plasma seed treatment improves chilling resistance of tomato plants through hydrogen peroxide and abscisic acid signaling pathway.Free Radic Biol Med. 2021 Aug 20;172:286-297. doi: 10.1016/j.freeradbiomed.2021.06.011. Epub 2021 Jun 15. Free Radic Biol Med. 2021. PMID: 34139310
Cited by
-
Light quality regulates plant biomass and fruit quality through a photoreceptor-dependent HY5-LHC/CYCB module in tomato.Hortic Res. 2023 Nov 16;10(12):uhad219. doi: 10.1093/hr/uhad219. eCollection 2023 Dec. Hortic Res. 2023. PMID: 38077493 Free PMC article.
-
Recent Insights into the Physio-Biochemical and Molecular Mechanisms of Low Temperature Stress in Tomato.Plants (Basel). 2024 Sep 28;13(19):2715. doi: 10.3390/plants13192715. Plants (Basel). 2024. PMID: 39409585 Free PMC article. Review.
-
Combining genome-wide association and genomic prediction to unravel the genetic architecture of carotenoid accumulation in carrot.Plant Genome. 2025 Mar;18(1):e20560. doi: 10.1002/tpg2.20560. Plant Genome. 2025. PMID: 39887573 Free PMC article.
-
Molecular breeding of tomato: Advances and challenges.J Integr Plant Biol. 2025 Mar;67(3):669-721. doi: 10.1111/jipb.13879. Epub 2025 Mar 18. J Integr Plant Biol. 2025. PMID: 40098531 Free PMC article. Review.
-
Physiological and transcriptomic comparisons shed light on the cold stress response mechanisms of Dendrobium spp.BMC Plant Biol. 2024 Apr 1;24(1):230. doi: 10.1186/s12870-024-04903-1. BMC Plant Biol. 2024. PMID: 38561687 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

