Reducing the number of accepted species in Aspergillus series Nigri
- PMID: 36760462
- PMCID: PMC9903907
- DOI: 10.3114/sim.2022.102.03
Reducing the number of accepted species in Aspergillus series Nigri
Abstract
The Aspergillus series Nigri contains biotechnologically and medically important species. They can produce hazardous mycotoxins, which is relevant due to the frequent occurrence of these species on foodstuffs and in the indoor environment. The taxonomy of the series has undergone numerous rearrangements, and currently, there are 14 species accepted in the series, most of which are considered cryptic. Species-level identifications are, however, problematic or impossible for many isolates even when using DNA sequencing or MALDI-TOF mass spectrometry, indicating a possible problem in the definition of species limits or the presence of undescribed species diversity. To re-examine the species boundaries, we collected DNA sequences from three phylogenetic markers (benA, CaM and RPB2) for 276 strains from series Nigri and generated 18 new whole-genome sequences. With the three-gene dataset, we employed phylogenetic methods based on the multispecies coalescence model, including four single-locus methods (GMYC, bGMYC, PTP and bPTP) and one multilocus method (STACEY). From a total of 15 methods and their various settings, 11 supported the recognition of only three species corresponding to the three main phylogenetic lineages: A. niger, A. tubingensis and A. brasiliensis. Similarly, recognition of these three species was supported by the GCPSR approach (Genealogical Concordance Phylogenetic Species Recognition) and analysis in DELINEATE software. We also showed that the phylogeny based on benA, CaM and RPB2 is suboptimal and displays significant differences from a phylogeny constructed using 5 752 single-copy orthologous proteins; therefore, the results of the delimitation methods may be subject to a higher than usual level of uncertainty. To overcome this, we randomly selected 200 genes from these genomes and performed ten independent STACEY analyses, each with 20 genes. All analyses supported the recognition of only one species in the A. niger and A. brasiliensis lineages, while one to four species were inconsistently delimited in the A. tubingensis lineage. After considering all of these results and their practical implications, we propose that the revised series Nigri includes six species: A. brasiliensis, A. eucalypticola, A. luchuensis (syn. A. piperis), A. niger (syn. A. vinaceus and A. welwitschiae), A. tubingensis (syn. A. chiangmaiensis, A. costaricensis, A. neoniger and A. pseudopiperis) and A. vadensis. We also showed that the intraspecific genetic variability in the redefined A. niger and A. tubingensis does not deviate from that commonly found in other aspergilli. We supplemented the study with a list of accepted species, synonyms and unresolved names, some of which may threaten the stability of the current taxonomy. Citation: Bian C, Kusuya Y, Sklenář F, D'hooge E, Yaguchi T, Ban S, Visagie CM, Houbraken J, Takahashi H, Hubka V (2022). Reducing the number of accepted species in Aspergillus series Nigri. Studies in Mycology 102: 95-132. doi: 10.3114/sim.2022.102.03.
Keywords: Aspergillus luchuensis; Aspergillus niger; Aspergillus tubingensis; clinical fungi; indoor fungi; infraspecific variability; multigene phylogeny, multispecies coalescence model; ochratoxin A; species delimitation.
© 2022 Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


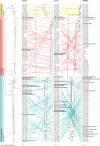
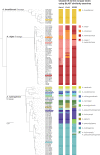



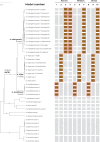



Similar articles
-
Taxonomy of Aspergillus series Versicolores: species reduction and lessons learned about intraspecific variability.Stud Mycol. 2022 Dec;102:53-93. doi: 10.3114/sim.2022.102.02. Epub 2022 Nov 16. Stud Mycol. 2022. PMID: 36760461 Free PMC article.
-
Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti.Stud Mycol. 2017 Sep;88:161-236. doi: 10.1016/j.simyco.2017.09.002. Epub 2017 Sep 27. Stud Mycol. 2017. PMID: 29158611 Free PMC article.
-
A monograph of Aspergillus section Candidi.Stud Mycol. 2022 Dec;102:1-51. doi: 10.3114/sim.2022.102.01. Epub 2022 Oct 19. Stud Mycol. 2022. PMID: 36760463 Free PMC article.
-
Review of secondary metabolites and mycotoxins from the Aspergillus niger group.Anal Bioanal Chem. 2009 Nov;395(5):1225-42. doi: 10.1007/s00216-009-3081-5. Epub 2009 Sep 16. Anal Bioanal Chem. 2009. PMID: 19756540 Review.
-
Taxonomy and significance of black aspergilli.Antonie Van Leeuwenhoek. 2004 Jul;86(1):33-49. doi: 10.1023/B:ANTO.0000024907.85688.05. Antonie Van Leeuwenhoek. 2004. PMID: 15103236 Review.
Cited by
-
A conceptual framework for nomenclatural stability and validity of medically important fungi: a proposed global consensus guideline for fungal name changes supported by ABP, ASM, CLSI, ECMM, ESCMID-EFISG, EUCAST-AFST, FDLC, IDSA, ISHAM, MMSA, and MSGERC.J Clin Microbiol. 2023 Nov 21;61(11):e0087323. doi: 10.1128/jcm.00873-23. Epub 2023 Oct 26. J Clin Microbiol. 2023. PMID: 37882528 Free PMC article.
-
Aspergillus latus: A cryptic causative agent of aspergillosis emerging in Japan.Med Mycol. 2025 Jun 5;63(6):myaf052. doi: 10.1093/mmy/myaf052. Med Mycol. 2025. PMID: 40493449 Free PMC article.
-
Combined Toxicity of the Most Common Indoor Aspergilli.Pathogens. 2023 Mar 14;12(3):459. doi: 10.3390/pathogens12030459. Pathogens. 2023. PMID: 36986381 Free PMC article.
-
Defining the relationship between phylogeny, clinical manifestation, and phenotype for Trichophyton mentagrophytes/interdigitale complex; a literature review and taxonomic recommendations.Med Mycol. 2023 May 1;61(5):myad042. doi: 10.1093/mmy/myad042. Med Mycol. 2023. PMID: 37070928 Free PMC article. Review.
-
Overview and Diversity of Fungi of the Genus Aspergillus Section Nigri on Maize and Small Grains.Foods. 2025 Jun 19;14(12):2146. doi: 10.3390/foods14122146. Foods. 2025. PMID: 40565755 Free PMC article.
References
-
- Ahrens D, Fujisawa T, Krammer H-J, et al. (2016). Rarity and incomplete sampling in DNA-based species delimitation. Systematic Biology 65: 478–494. - PubMed
-
- Al-Musallam (1980). A revision of the black Aspergillus species. Ph.D. dissertation. Rijksuniversiteit; Utrecht, Netherlands.
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
