Synthetic lethal interactions of DEAD/H-box helicases as targets for cancer therapy
- PMID: 36761420
- PMCID: PMC9905851
- DOI: 10.3389/fonc.2022.1087989
Synthetic lethal interactions of DEAD/H-box helicases as targets for cancer therapy
Abstract
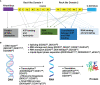
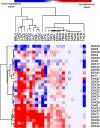
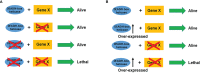
DEAD/H-box helicases are implicated in virtually every aspect of RNA metabolism, including transcription, pre-mRNA splicing, ribosomes biogenesis, nuclear export, translation initiation, RNA degradation, and mRNA editing. Most of these helicases are upregulated in various cancers and mutations in some of them are associated with several malignancies. Lately, synthetic lethality (SL) and synthetic dosage lethality (SDL) approaches, where genetic interactions of cancer-related genes are exploited as therapeutic targets, are emerging as a leading area of cancer research. Several DEAD/H-box helicases, including DDX3, DDX9 (Dbp9), DDX10 (Dbp4), DDX11 (ChlR1), and DDX41 (Sacy-1), have been subjected to SL analyses in humans and different model organisms. It remains to be explored whether SDL can be utilized to identity druggable targets in DEAD/H-box helicase overexpressing cancers. In this review, we analyze gene expression data of a subset of DEAD/H-box helicases in multiple cancer types and discuss how their SL/SDL interactions can be used for therapeutic purposes. We also summarize the latest developments in clinical applications, apart from discussing some of the challenges in drug discovery in the context of targeting DEAD/H-box helicases.
Keywords: DEAD/H-box helicase; cancer; drug development; synthetic dosage lethality; synthetic lethality; therapeutic target.
Copyright © 2023 Arna, Patel, Singh, Vizeacoumar, Kusalik, Freywald, Vizeacoumar and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
DDX41: exploring the roles of a versatile helicase.Biochem Soc Trans. 2024 Feb 28;52(1):395-405. doi: 10.1042/BST20230725. Biochem Soc Trans. 2024. PMID: 38348889 Free PMC article. Review.
-
DEAD-box helicase family proteins: emerging targets in digestive system cancers and advances in targeted drug development.J Transl Med. 2024 Dec 20;22(1):1120. doi: 10.1186/s12967-024-05930-0. J Transl Med. 2024. PMID: 39707322 Free PMC article. Review.
-
DEAD/H-box helicases:Anti-viral and pro-viral roles during infections.Virus Res. 2022 Feb;309:198658. doi: 10.1016/j.virusres.2021.198658. Epub 2021 Dec 17. Virus Res. 2022. PMID: 34929216 Review.
-
Insights into the Involvement of Spliceosomal Mutations in Myelodysplastic Disorders from Analysis of SACY-1/DDX41 in Caenorhabditis elegans.Genetics. 2020 Apr;214(4):869-893. doi: 10.1534/genetics.119.302973. Epub 2020 Feb 14. Genetics. 2020. PMID: 32060018 Free PMC article.
-
DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs.Molecules. 2020 Feb 24;25(4):1015. doi: 10.3390/molecules25041015. Molecules. 2020. PMID: 32102413 Free PMC article. Review.
Cited by
-
Spliceosomal GTPase EFTUD2 mediates DDX41 intron retention to promote the malignant progression of ovarian cancer.Br J Cancer. 2025 Sep;133(4):508-523. doi: 10.1038/s41416-025-03079-1. Epub 2025 Jun 24. Br J Cancer. 2025. PMID: 40555777
-
DDX41: exploring the roles of a versatile helicase.Biochem Soc Trans. 2024 Feb 28;52(1):395-405. doi: 10.1042/BST20230725. Biochem Soc Trans. 2024. PMID: 38348889 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

