Vascular inflammation on a chip: A scalable platform for trans-endothelial electrical resistance and immune cell migration
- PMID: 36761747
- PMCID: PMC9903066
- DOI: 10.3389/fimmu.2023.1118624
Vascular inflammation on a chip: A scalable platform for trans-endothelial electrical resistance and immune cell migration
Abstract
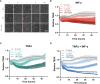
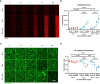
The vasculature system plays a critical role in inflammation processes in the body. Vascular inflammatory mechanisms are characterized by disruption of blood vessel wall permeability together with increased immune cell recruitment and migration. There is a critical need to develop models that fully recapitulate changes in vascular barrier permeability in response to inflammatory conditions. We developed a scalable platform for parallel measurements of trans epithelial electrical resistance (TEER) in 64 perfused microfluidic HUVEC tubules under inflammatory conditions. Over 250 tubules where exposed to Tumor necrosis factor alpha (TNFα) and interferon gamma (INF-γ) or human peripheral blood mononuclear cells. The inflammatory response was quantified based on changes TEER and expression of ICAM and VE-cadherin. We observed changes in barrier function in the presence of both inflammatory cytokines and human peripheral blood mononuclear cells, characterized by decreased TEER values, increase in ICAM expression as well changes in endothelial morphology. OrganoPlate 3-lane64 based HUVEC tubules provide a valuable tool for inflammatory studies in an automation compatible manner. Continuous TEER measurements enable long term, sensitive assays for barrier studies. We propose the use of our platform as a powerful tool for modelling endothelial inflammation in combination with immune cell interaction that can be used to screen targets and drugs to treat chronic vascular inflammation.
Keywords: TEER; TNFα, INF-γ and OrganoPlate; endothelium; immune cell migration; organ-on-a-chip; vascular inflammation.
Copyright © 2023 Ehlers, Nicolas, Schavemaker, Heijmans, Bulst, Trietsch and van den Broek.
Conflict of interest statement
HE, AN, FS, ST, and LB are employees of Mimetas B.V., which markets the OrganoPlate, OrganoTEER and OrganoFlow, and holds the registered trademarks OrganoPlate, OrganoTEER and OrganoFlow. ST is shareholder of Mimetas B.V. MB is employed by Sciospec GmbH.
Figures



References
-
- Pahwa R, Goyal A, Jialal I. Chronic inflammation. In: Pathobiol hum dis a dyn encycl dis mech (2022). [Book StatPearls: StatPearls Publishing, Treasure Island (FL)] Available at: https://www.ncbi.nlm.nih.gov/books/NBK493173/. - PubMed
-
- Zanoli L, Briet M, Empana JP, Guimarães Cunha P, Mäki-Petäjä M, Protogerou AD, et al. . Vascular consequences of inflammation: A position statement from the ESH working group on vascular structure and function and the ARTERY society. J Hypertens (2020) 38(9):1682–98. doi: 10.1097/HJH.0000000000002508 - DOI - PMC - PubMed
-
- Cardiovascular diseases . Available at: https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

