Macrophage differentiation is marked by increased abundance of the mRNA 3' end processing machinery, altered poly(A) site usage, and sensitivity to the level of CstF64
- PMID: 36761770
- PMCID: PMC9905730
- DOI: 10.3389/fimmu.2023.1091403
Macrophage differentiation is marked by increased abundance of the mRNA 3' end processing machinery, altered poly(A) site usage, and sensitivity to the level of CstF64
Abstract
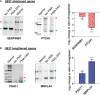
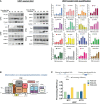
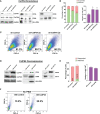
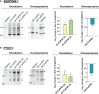
Regulation of mRNA polyadenylation is important for response to external signals and differentiation in several cell types, and results in mRNA isoforms that vary in the amount of coding sequence or 3' UTR regulatory elements. However, its role in differentiation of monocytes to macrophages has not been investigated. Macrophages are key effectors of the innate immune system that help control infection and promote tissue-repair. However, overactivity of macrophages contributes to pathogenesis of many diseases. In this study, we show that macrophage differentiation is characterized by shortening and lengthening of mRNAs in relevant cellular pathways. The cleavage/polyadenylation (C/P) proteins increase during differentiation, suggesting a possible mechanism for the observed changes in poly(A) site usage. This was surprising since higher C/P protein levels correlate with higher proliferation rates in other systems, but monocytes stop dividing after induction of differentiation. Depletion of CstF64, a C/P protein and known regulator of polyadenylation efficiency, delayed macrophage marker expression, cell cycle exit, attachment, and acquisition of structural complexity, and impeded shortening of mRNAs with functions relevant to macrophage biology. Conversely, CstF64 overexpression increased use of promoter-proximal poly(A) sites and caused the appearance of differentiated phenotypes in the absence of induction. Our findings indicate that regulation of polyadenylation plays an important role in macrophage differentiation.
Keywords: CSTF2; CstF64; alternative polyadenylation; differentiation; macrophage; monocyte.
Copyright © 2023 Mukherjee, Graber and Moore.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Cleavage stimulating factor 64 depletion mitigates cardiac fibrosis through alternative polyadenylation.Biochem Biophys Res Commun. 2022 Mar 15;597:109-114. doi: 10.1016/j.bbrc.2022.01.093. Epub 2022 Jan 29. Biochem Biophys Res Commun. 2022. PMID: 35134608 Free PMC article.
-
Implications of polyadenylation in health and disease.Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
-
Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3' processing.RNA. 2013 Dec;19(12):1781-90. doi: 10.1261/rna.042317.113. Epub 2013 Oct 22. RNA. 2013. PMID: 24149845 Free PMC article.
-
Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18773-8. doi: 10.1073/pnas.1211101109. Epub 2012 Oct 29. Proc Natl Acad Sci U S A. 2012. PMID: 23112178 Free PMC article.
-
Poly(A) tale: From A to A; RNA polyadenylation in prokaryotes and eukaryotes.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1837. doi: 10.1002/wrna.1837. Wiley Interdiscip Rev RNA. 2024. PMID: 38485452 Review.
Cited by
-
The emerging theme of 3'UTR mRNA isoform regulation in reprogramming of cell metabolism.Biochem Soc Trans. 2023 Jun 28;51(3):1111-1119. doi: 10.1042/BST20221128. Biochem Soc Trans. 2023. PMID: 37171086 Free PMC article. Review.
-
Mycobacterium tuberculosis modulates NUDT21-mediated alternative polyadenylation to enhance FTH1 expression in macrophages and promotes intracellular growth.Front Cell Infect Microbiol. 2025 May 2;15:1578163. doi: 10.3389/fcimb.2025.1578163. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40384984 Free PMC article.
-
RNA processing in innate immunity: regulation by RNA-binding proteins.Trends Biochem Sci. 2025 Jul;50(7):610-621. doi: 10.1016/j.tibs.2025.04.004. Epub 2025 May 15. Trends Biochem Sci. 2025. PMID: 40379525 Review.
-
Downregulation of CPSF6 leads to global mRNA 3' UTR shortening and enhanced antiviral immune responses.PLoS Pathog. 2024 Feb 28;20(2):e1012061. doi: 10.1371/journal.ppat.1012061. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38416782 Free PMC article.
-
Elevated pre-mRNA 3' end processing activity in cancer cells renders vulnerability to inhibition of cleavage and polyadenylation.Nat Commun. 2023 Aug 1;14(1):4480. doi: 10.1038/s41467-023-39793-8. Nat Commun. 2023. PMID: 37528120 Free PMC article.
References
-
- Mammana S, Fagone P, Cavalli E, Basile MS, Petralia MC, Nicoletti F, et al. . The role of macrophages in neuroinflammatory and neurodegenerative pathways of alzheimer's disease, amyotrophic lateral sclerosis, and multiple sclerosis: Pathogenetic cellular effectors and potential therapeutic targets. Int J Mol Sci (2018) 19(3). doi: 10.3390/ijms19030831 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

