A comparison of gene organisations and phylogenetic relationships of all 22 squamate species listed in South Korea using complete mitochondrial DNA
- PMID: 36761844
- PMCID: PMC9836557
- DOI: 10.3897/zookeys.1129.82981
A comparison of gene organisations and phylogenetic relationships of all 22 squamate species listed in South Korea using complete mitochondrial DNA
Abstract
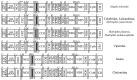
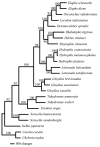
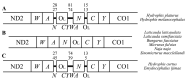
Studies using complete mitochondrial genome data have the potential to increase our understanding on gene organisations and evolutionary species relationships. In this study, we compared complete mitochondrial genomes between all 22 squamate species listed in South Korea. In addition, we constructed Maximum Parsimony (MP), Maximum Likelihood (ML) and Bayesian Inference (BI) phylogenetic trees using 13 mitochondrial protein-coding genes. The mitochondrial genes for all six species in the suborder Sauria followed the same organisation as the sequenced Testudines (turtle) outgroup. In contrast, 16 snake species in the suborder Serpentes contained some gene organisational variations. For example, all snake species contained a second control region (CR2), while three species in the family Viperidae had a translocated tRNA-Pro gene region. In addition, the snake species, Elapheschrenckii, carried a tRNA-Pro pseudogene. We were also able to identify a translocation of a tRNA-Asn gene within the five tRNA (WANCY gene region) gene clusters for two true sea snake species in the subfamily Hydrophiinae. Our BI phylogenetic tree was also well fitted against currently known Korean squamate phylogenetic trees, where each family and genus unit forms monophyletic clades and the suborder Sauria is paraphyletic to the suborder Serpentes. Our results may form the basis for future northeast Asian squamate phylogenetic studies.
Keywords: Full mitochondrial genome; Korea; Squamata; WANCY; phylogeny; rearrangement; tRNA-Pro.
Daesik Park, Il-Hun Kim, Il-Kook Park, Alejandro Grajal-Puche, Jaejin Park.
Figures

References
-
- An JH, Park DS, Lee JH, Kim KS, Lee H, Min MS. (2010) No genetic differentiation of Elapheschrenckii subspecies in Korea based on 9 microsatellite loci. Animal Systematics, Evolution and Diversity 26(1): 15–19. 10.5635/KJSZ.2010.26.1.015 - DOI
-
- Boore JL. (2000) The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. In: Sankoff D, Nadezu JH. (Eds) Comparative genomics.Springer Press, Dordrecht, 133–147. 10.1007/978-94-011-4309-7_13 - DOI
LinkOut - more resources
Full Text Sources
