A DNA barcode library for katydids, cave crickets, and leaf-rolling crickets (Tettigoniidae, Rhaphidophoridae and Gryllacrididae) from Zhejiang Province, China
- PMID: 36762040
- PMCID: PMC9836636
- DOI: 10.3897/zookeys.1123.86704
A DNA barcode library for katydids, cave crickets, and leaf-rolling crickets (Tettigoniidae, Rhaphidophoridae and Gryllacrididae) from Zhejiang Province, China
Abstract
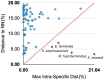
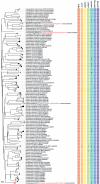
Barcode libraries are generally assembled with two main objectives in mind: specimen identification and species discovery/delimitation. In this study, the standard COI barcode region was sequenced from 681 specimens belonging to katydids (Tettigoniidae), cave crickets (Rhaphidophoridae), and leaf-rolling crickets (Gryllacrididae) from Zhejiang Province, China. Of these, four COI-5P sequences were excluded from subsequent analyses because they were likely NUMTs (nuclear mitochondrial pseudogenes). The final dataset consisted of 677 barcode sequences representing 90 putative species-level taxa. Automated cluster delineation using the Barcode of Life Data System (BOLD) revealed 118 BINs (Barcodes Index Numbers). Among these 90 species-level taxa, 68 corresponded with morphospecies, while the remaining 22 were identified based on reverse taxonomy using BIN assignment. Thirteen of these morphospecies were represented by a single barcode (so-called singletons), and each of 19 morphospecies were split into more than one BIN. The consensus delimitation scheme yielded 55 Molecular Operational Taxonomic Units (MOTUs). Only four morphospecies (I max > DNN) failed to be recovered as monophyletic clades (i.e., Elimaeaterminalis, Phyllomimusklapperichi, Sinochloraszechwanensis and Xizicushowardi), so it is speculated that these may be species complexes. Therefore, the diversity of katydids, cave crickets, and leaf-rolling crickets in Zhejiang Province is probably slightly higher than what current taxonomy would suggest.
Keywords: Barcode Index Number; Ensifera; Orthoptera; cryptic species; species delimitation.
Yizheng Zhao, Hui Wang, Huimin Huang, Zhijun Zhou.
Figures



References
-
- Ahrens D, Fujisawa T, Krammer HJ, Eberle J, Fabrizi S, Vogler AP. (2016) Rarity and incomplete sampling in DNA-based species delimitation. Systematic Biology 65(3): 478–494. http://10.1093/sysbio/syw002 - DOI - PubMed
-
- Andrew R. (2016) FigTree: Tree figure drawing tool Version 1.4.3. Institute of Evolutionary Biology, United Kingdom, University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/
LinkOut - more resources
Full Text Sources
