Transcription factor ZBTB42 is a novel prognostic factor associated with immune cell infiltration in glioma
- PMID: 36762114
- PMCID: PMC9905726
- DOI: 10.3389/fphar.2023.1102277
Transcription factor ZBTB42 is a novel prognostic factor associated with immune cell infiltration in glioma
Abstract
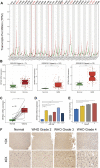
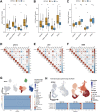
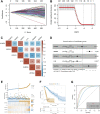
Background: ZBTB42 is a transcription factor that belongs to the ZBTB transcript factor family and plays an important role in skeletal muscle development. Dysregulation of ZBTB42 expression can lead to a variety of diseases. However, the function of ZBTB42 in glioma development has not been studied by now. Methods: We analyzed the expression of ZBTB42 in LGG and GBM via the The Cancer Genome Atlas CGA and Chinese Glioma Genome Atlas database. Gene Ontology, KEGG, and GSVA analyses were performed to illustrate ZBTB42-related pathways. ESTIMATE and CIBERSORT were applied to calculate the immune score and immune cell proportion in glioma. One-class logistic regression OCLR algorithm was used to study the stemness of glioma. Multivariate Cox analysis was employed to detect the prognostic value of five ZBTB42-related genes. Results: Our results show that ZBTB42 is highly expressed in glioma and may be a promising prognostic factor for Low Grade Glioma and GBM. In addition, ZBTB42 is related to immune cell infiltration and may play a role in the immune suppression microenvironment. What's more, ZBTB42 is correlated with stem cell markers and positively associated with glioma stemness. Finally, a five genes nomogram based on ZBTB42 was constructed and has an effective prognosis prediction ability. Conclusion: We identify that ZBTB42 is a prognostic biomarker for Low Grade Glioma and GBM and its function is related to the suppressive tumor microenvironment and stemness of glioma.
Keywords: ZBTB42; glioma; immune suppression; microenvironment; stemness.
Copyright © 2023 Li, Zhu, Chen, Xia, Adegboro, Wanggou and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Choi W. I., Kim M. Y., Jeon B. N., Koh D. I., Yun C. O., Li Y., et al. (2014). Role of promyelocytic leukemia zinc finger (PLZF) in cell proliferation and cyclin-dependent kinase inhibitor 1A (p21WAF/CDKN1A) gene repression. J. Biol. Chem. 289, 18625–18640. 10.1074/jbc.M113.538751 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

