Catalytic deAMPylation in AMPylation-inhibitory/assistant forms of FICD protein
- PMID: 36762200
- PMCID: PMC9905249
- DOI: 10.3389/fchem.2023.1077188
Catalytic deAMPylation in AMPylation-inhibitory/assistant forms of FICD protein
Abstract
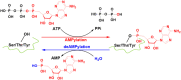
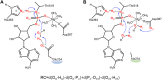
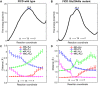
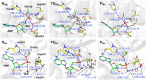
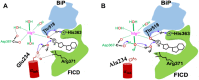
DeAMPylation, as a reversible reaction of AMPylation and mediated by the endoplasmic reticulum-localized enzyme FICD (filamentation induced by cAMP domain protein, also known as HYPE), is an important process in protein posttranslational modifications (PTMs). Elucidating the function and catalytic details of FICD is of vital importance to provide a comprehensive understanding of protein folding homeostasis. However, the detailed deAMPylation mechanism is still unclear. Furthermore, the role of a conserved glutamine (Glu234), that plays an inhibitory role in the AMPylation response, is still an open question in the deAMPylation process. In the present work, the elaborated deAMPylation mechanisms with AMPylation-inhibitory/assistant forms of FICD (wild type and Glu234Ala mutant) were investigated based on the QM(DFT)/MM MD approach. The results revealed that deAMPylation was triggered by proton transfer from protonated histidine (His363) to AMPylated threonine, instead of a nucleophilic attack of water molecules adding to the phosphorus of AMP. The free energy barrier of deAMPylation in the wild type (∼17.3 kcal/mol) is consistent with that in the Glu234Ala mutant of FICD (∼17.1 kcal/mol), suggesting that the alteration of the Glu234 residue does not affect the deAMPylation reaction and indirectly verifying the inducement of deAMPylation in FICD. In the wild type, the proton in the nucleophilic water molecule is transferred to Glu234, whereas it is delivered to Asp367 through the hydrogen-bond network of coordinated water molecules in the Glu234Ala mutant. The present findings were inspirational for understanding the catalytic and inhibitory mechanisms of FICD-mediated AMP transfer, paving the way for further studies on the physiological role of FICD protein.
Keywords: FICD protein; QM/MM MD; catalytic mechanism; deAMPylation; inhibitory helix.
Copyright © 2023 Liu, Li, Wang, Wang and Tang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Investigation of the Detailed AMPylated Reaction Mechanism for the Huntingtin Yeast-Interacting Protein E Enzyme HYPE.Int J Mol Sci. 2021 Jun 29;22(13):6999. doi: 10.3390/ijms22136999. Int J Mol Sci. 2021. PMID: 34209803 Free PMC article.
-
Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD.Nat Commun. 2021 Aug 18;12(1):5004. doi: 10.1038/s41467-021-25076-7. Nat Commun. 2021. PMID: 34408154 Free PMC article.
-
An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.EMBO J. 2019 Oct 4;38(21):e102177. doi: 10.15252/embj.2019102177. Epub 2019 Sep 18. EMBO J. 2019. PMID: 31531998 Free PMC article.
-
From Young to Old: AMPylation Hits the Brain.Cell Chem Biol. 2020 Jul 16;27(7):773-779. doi: 10.1016/j.chembiol.2020.05.009. Epub 2020 Jun 9. Cell Chem Biol. 2020. PMID: 32521229 Review.
-
Enzymes Involved in AMPylation and deAMPylation.Chem Rev. 2018 Feb 14;118(3):1199-1215. doi: 10.1021/acs.chemrev.7b00145. Epub 2017 Aug 18. Chem Rev. 2018. PMID: 28819965 Free PMC article. Review.
References
-
- Bayly C. I., Cieplak P., Cornell W., Kollman P. A. (1993). A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges:the RESP model. The Journal of Physical Chemistry. 97 (40), 10269–10280. 10.1021/j100142a004 - DOI
-
- Beeman D. (1976). Some multistep methods for use in molecular dynamics calculations. The Journal of Physical Chemistry. 20, 130–139. 10.1016/0021-9991(76)90059-0 - DOI
-
- Brown M. S., Segal A., Stadtman E. R. (1971). Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the P II -regulatory protein. Proceedings of the National Academy of Sciences of the United States of America. 68, 2949–2953. 10.1073/pnas.68.12.2949 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

