The Raf-like MAPKKK INTEGRIN-LINKED KINASE 5 regulates purinergic receptor-mediated innate immunity in Arabidopsis
- PMID: 36762404
- PMCID: PMC10118279
- DOI: 10.1093/plcell/koad029
The Raf-like MAPKKK INTEGRIN-LINKED KINASE 5 regulates purinergic receptor-mediated innate immunity in Arabidopsis
Abstract
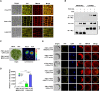
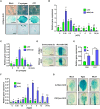
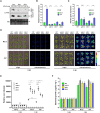
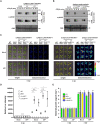
Mitogen-activated protein (MAP) kinase signaling cascades play important roles in eukaryotic defense against various pathogens. Activation of the extracellular ATP (eATP) receptor P2K1 triggers MAP kinase 3 and 6 (MPK3/6) phosphorylation, which leads to an elevated plant defense response. However, the mechanism by which P2K1 activates the MAPK cascade is unclear. In this study, we show that in Arabidopsis thaliana, P2K1 phosphorylates the Raf-like MAP kinase kinase kinase (MAPKKK) INTEGRIN-LINKED KINASE 5 (ILK5) on serine 192 in the presence of eATP. The interaction between P2K1 and ILK5 was confirmed both in vitro and in planta and their interaction was enhanced by ATP treatment. Similar to P2K1 expression, ILK5 expression levels were highly induced by treatment with ATP, flg22, Pseudomonas syringae pv. tomato DC3000, and various abiotic stresses. ILK5 interacts with and phosphorylates the MAP kinase MKK5. Moreover, phosphorylation of MPK3/6 was significantly reduced upon ATP treatment in ilk5 mutant plants, relative to wild-type (WT). The ilk5 mutant plants showed higher susceptibility to P. syringae pathogen infection relative to WT plants. Plants expressing only the mutant ILK5S192A protein, with decreased kinase activity, did not activate the MAPK cascade upon ATP addition. These results suggest that eATP activation of P2K1 results in transphosphorylation of the Raf-like MAPKKK ILK5, which subsequently triggers the MAPK cascade, culminating in activation of MPK3/6 associated with an elevated innate immune response.
© American Society of Plant Biologists 2023. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare no conflict of interest.
Figures



Similar articles
-
Phosphorylation-mediated regulation of integrin-linked kinase 5 by purinoreceptor P2K2.Plant Signal Behav. 2023 Dec 31;18(1):2261743. doi: 10.1080/15592324.2023.2261743. Epub 2023 Dec 17. Plant Signal Behav. 2023. PMID: 37750411 Free PMC article.
-
Arabidopsis Lectin Receptor Kinase P2K2 Is a Second Plant Receptor for Extracellular ATP and Contributes to Innate Immunity.Plant Physiol. 2020 Jul;183(3):1364-1375. doi: 10.1104/pp.19.01265. Epub 2020 Apr 28. Plant Physiol. 2020. PMID: 32345768 Free PMC article.
-
EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity.PLoS Genet. 2014 May 15;10(5):e1004389. doi: 10.1371/journal.pgen.1004389. eCollection 2014. PLoS Genet. 2014. PMID: 24830651 Free PMC article.
-
Recent advances in understanding the role of two mitogen-activated protein kinase cascades in plant immunity.J Exp Bot. 2024 Apr 15;75(8):2256-2265. doi: 10.1093/jxb/erae020. J Exp Bot. 2024. PMID: 38241698 Review.
-
ATP homeostasis and signaling in plants.Plant Commun. 2024 Apr 8;5(4):100834. doi: 10.1016/j.xplc.2024.100834. Epub 2024 Feb 7. Plant Commun. 2024. PMID: 38327057 Free PMC article. Review.
Cited by
-
Identifying Receptor Kinase Substrates Using an 8000 Peptide Kinase Client Library Enriched for Conserved Phosphorylation Sites.Mol Cell Proteomics. 2025 Mar;24(3):100926. doi: 10.1016/j.mcpro.2025.100926. Epub 2025 Feb 7. Mol Cell Proteomics. 2025. PMID: 39923935 Free PMC article.
-
Unveiling orphan receptor-like kinases in plants: novel client discovery using high-confidence library predictions in the Kinase-Client (KiC) assay.Front Plant Sci. 2024 Apr 3;15:1372361. doi: 10.3389/fpls.2024.1372361. eCollection 2024. Front Plant Sci. 2024. PMID: 38633461 Free PMC article.
-
Identification of mitogen-activated protein kinases substrates in Arabidopsis using kinase client assay.Plant Signal Behav. 2024 Dec 31;19(1):2326238. doi: 10.1080/15592324.2024.2326238. Epub 2024 Mar 17. Plant Signal Behav. 2024. PMID: 38493505 Free PMC article.
-
Salt stress releases extracellular ATP to activate purinergic signaling and inhibit plant growth.Plant Physiol. 2023 Oct 26;193(3):1753-1757. doi: 10.1093/plphys/kiad429. Plant Physiol. 2023. PMID: 37506300 Free PMC article.
-
Extracellular ATP: an emerging multifaceted regulator of plant fitness.Plant Biotechnol J. 2025 May;23(5):1771-1782. doi: 10.1111/pbi.70006. Epub 2025 Feb 12. Plant Biotechnol J. 2025. PMID: 39937654 Free PMC article. Review.
References
-
- Balagué C, Gouget A, Bouchez O, Souriac C, Haget N, Boutet-Mercey S, Govers F, Roby D, Canut H. The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol Plant Pathol. 2017:18(7):937–948. 10.1111/mpp.12457 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

