Transcriptional pause extension benefits the stand-by rather than catch-up Rho-dependent termination
- PMID: 36762473
- PMCID: PMC10085680
- DOI: 10.1093/nar/gkad051
Transcriptional pause extension benefits the stand-by rather than catch-up Rho-dependent termination
Abstract
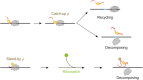
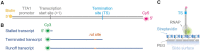
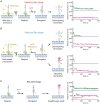
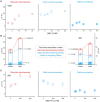
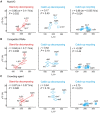
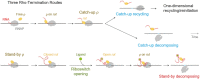
Transcriptional pause is essential for all types of termination. In this single-molecule study on bacterial Rho factor-dependent terminators, we confirm that the three Rho-dependent termination routes operate compatibly together in a single terminator, and discover that their termination efficiencies depend on the terminational pauses in unexpected ways. Evidently, the most abundant route is that Rho binds nascent RNA first and catches up with paused RNA polymerase (RNAP) and this catch-up Rho mediates simultaneous releases of transcript RNA and template DNA from RNAP. The fastest route is that the catch-up Rho effects RNA-only release and leads to 1D recycling of RNAP on DNA. The slowest route is that the RNAP-prebound stand-by Rho facilitates only the simultaneous rather than sequential releases. Among the three routes, only the stand-by Rho's termination efficiency positively correlates with pause duration, contrary to a long-standing speculation, invariably in the absence or presence of NusA/NusG factors, competitor RNAs or a crowding agent. Accordingly, the essential terminational pause does not need to be long for the catch-up Rho's terminations, and long pauses benefit only the stand-by Rho's terminations. Furthermore, the Rho-dependent termination of mgtA and ribB riboswitches is controlled mainly by modulation of the stand-by rather than catch-up termination.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Landick R. Transcriptional pausing as a mediator of bacterial gene regulation. Annu. Rev. Microbiol. 2021; 75:291–314. - PubMed
-
- Nickels B.E., Roberts C.W., Sun H., Roberts J.W., Hochschild A.. The σ70 subunit of RNA polymerase is contacted by the λ Q antiterminator during early elongation. Mol. Cell. 2002; 10:611–622. - PubMed
-
- Naftelberg S., Schor I.E., Ast G., Kornblihtt A.R.. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015; 84:165–198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

