Identification of a novel 5-aminomethyl-2-thiouridine methyltransferase in tRNA modification
- PMID: 36762482
- PMCID: PMC9976899
- DOI: 10.1093/nar/gkad048
Identification of a novel 5-aminomethyl-2-thiouridine methyltransferase in tRNA modification
Abstract
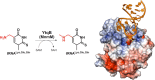
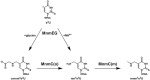
The uridine at the 34th position of tRNA, which is able to base pair with the 3'-end codon on mRNA, is usually modified to influence many aspects of decoding properties during translation. Derivatives of 5-methyluridine (xm5U), which include methylaminomethyl (mnm-) or carboxymethylaminomethyl (cmnm-) groups at C5 of uracil base, are widely conserved at the 34th position of many prokaryotic tRNAs. In Gram-negative bacteria such as Escherichia coli, a bifunctional MnmC is involved in the last two reactions of the biosynthesis of mnm5(s2)U, in which the enzyme first converts cmnm5(s2)U to 5-aminomethyl-(2-thio)uridine (nm5(s2)U) and subsequently installs the methyl group to complete the formation of mnm5(s2)U. Although mnm5s2U has been identified in tRNAs of Gram-positive bacteria and plants as well, their genomes do not contain an mnmC ortholog and the gene(s) responsible for this modification is unknown. We discovered that MnmM, previously known as YtqB, is the methyltransferase that converts nm5s2U to mnm5s2U in Bacillus subtilis through comparative genomics, gene complementation experiments, and in vitro assays. Furthermore, we determined X-ray crystal structures of MnmM complexed with anticodon stem loop of tRNAGln. The structures provide the molecular basis underlying the importance of U33-nm5s2U34-U35 as the key determinant for the specificity of MnmM.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Structural basis for hypermodification of the wobble uridine in tRNA by bifunctional enzyme MnmC.BMC Struct Biol. 2013 Apr 24;13:5. doi: 10.1186/1472-6807-13-5. BMC Struct Biol. 2013. PMID: 23617613 Free PMC article.
-
The output of the tRNA modification pathways controlled by the Escherichia coli MnmEG and MnmC enzymes depends on the growth conditions and the tRNA species.Nucleic Acids Res. 2014 Feb;42(4):2602-23. doi: 10.1093/nar/gkt1228. Epub 2013 Nov 30. Nucleic Acids Res. 2014. PMID: 24293650 Free PMC article.
-
Transfer RNA(5-methylaminomethyl-2-thiouridine)-methyltransferase from Escherichia coli K-12 has two enzymatic activities.J Biol Chem. 1987 Jun 25;262(18):8488-95. J Biol Chem. 1987. PMID: 3298234
-
Transfer RNA Modification Enzymes with a Thiouridine Synthetase, Methyltransferase and Pseudouridine Synthase (THUMP) Domain and the Nucleosides They Produce in tRNA.Genes (Basel). 2023 Jan 31;14(2):382. doi: 10.3390/genes14020382. Genes (Basel). 2023. PMID: 36833309 Free PMC article. Review.
-
Modification of the wobble uridine in bacterial and mitochondrial tRNAs reading NNA/NNG triplets of 2-codon boxes.RNA Biol. 2014;11(12):1495-507. doi: 10.4161/15476286.2014.992269. RNA Biol. 2014. PMID: 25607529 Free PMC article. Review.
Cited by
-
A tRNA modification in Mycobacterium tuberculosis facilitates optimal intracellular growth.bioRxiv [Preprint]. 2023 Jun 9:2023.02.20.529267. doi: 10.1101/2023.02.20.529267. bioRxiv. 2023. Update in: Elife. 2023 Sep 27;12:RP87146. doi: 10.7554/eLife.87146. PMID: 36865327 Free PMC article. Updated. Preprint.
-
RudS: bacterial desulfidase responsible for tRNA 4-thiouridine de-modification.Nucleic Acids Res. 2024 Sep 23;52(17):10543-10562. doi: 10.1093/nar/gkae716. Nucleic Acids Res. 2024. PMID: 39166491 Free PMC article.
-
A tRNA modification in Mycobacterium tuberculosis facilitates optimal intracellular growth.Elife. 2023 Sep 27;12:RP87146. doi: 10.7554/eLife.87146. Elife. 2023. PMID: 37755167 Free PMC article.
-
Alternate routes to mnm5s2U synthesis in Gram-positive bacteria.J Bacteriol. 2024 Apr 18;206(4):e0045223. doi: 10.1128/jb.00452-23. Epub 2024 Mar 29. J Bacteriol. 2024. PMID: 38551342 Free PMC article.
-
tRNA modifying enzymes MnmE and MnmG are essential for Plasmodium falciparum apicoplast maintenance.bioRxiv [Preprint]. 2025 Jan 6:2024.12.21.629855. doi: 10.1101/2024.12.21.629855. bioRxiv. 2025. PMID: 39763917 Free PMC article. Preprint.
References
-
- Soma A., Ikeuchi Y., Kanemasa S., Kobayashi K., Ogasawara N., Ote T., Kato J.I., Watanabe K., Sekine Y., Suzuki T.. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003; 12:689–698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

